Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
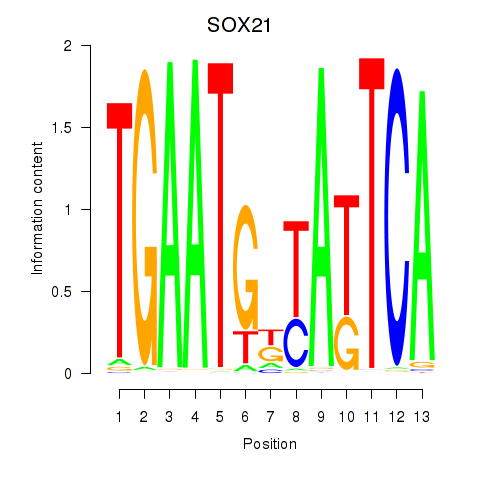
Results for SOX21
Z-value: 0.78
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SRY-box transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg19_v2_chr13_-_95364389_95364389 | -0.42 | 5.8e-01 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_42381173 | 0.62 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr12_+_1099675 | 0.47 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr19_+_42381337 | 0.46 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr7_-_107968999 | 0.42 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr18_-_47018869 | 0.42 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr15_-_80263506 | 0.36 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr2_-_241195452 | 0.34 |
ENST00000457178.1
|
AC124861.1
|
AC124861.1 |
| chr1_+_67632083 | 0.33 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr11_+_35201826 | 0.32 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_169455169 | 0.32 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr12_-_89746264 | 0.32 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr8_-_50466973 | 0.32 |
ENST00000520800.1
|
RP11-738G5.2
|
Uncharacterized protein |
| chr18_-_47018769 | 0.30 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr18_-_47018897 | 0.30 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr12_-_1058849 | 0.30 |
ENST00000358495.3
|
RAD52
|
RAD52 homolog (S. cerevisiae) |
| chr7_-_86849836 | 0.29 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr1_+_221051699 | 0.29 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr3_+_186288454 | 0.24 |
ENST00000265028.3
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr1_+_151171012 | 0.23 |
ENST00000349792.5
ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr6_+_131958436 | 0.22 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr5_-_55529115 | 0.20 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr14_-_102771462 | 0.20 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr6_+_131894284 | 0.20 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chrY_+_14813160 | 0.19 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr4_-_103746924 | 0.18 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103746683 | 0.17 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_131353461 | 0.17 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chrX_-_153210107 | 0.17 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr4_-_103747011 | 0.16 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_+_18682531 | 0.16 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chrX_-_55208866 | 0.16 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr5_-_95158644 | 0.16 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr12_+_59194154 | 0.15 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr14_+_64565442 | 0.14 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr19_-_45737469 | 0.14 |
ENST00000413988.1
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr14_+_60712463 | 0.13 |
ENST00000325642.3
ENST00000529574.1 |
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr12_-_10324716 | 0.13 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr17_+_60457914 | 0.13 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr3_+_155838337 | 0.13 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr7_+_6713376 | 0.12 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr19_-_2783255 | 0.11 |
ENST00000589251.1
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chrX_-_138724677 | 0.11 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr5_+_175875349 | 0.11 |
ENST00000261942.6
|
FAF2
|
Fas associated factor family member 2 |
| chr10_-_12084770 | 0.10 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr14_+_95048217 | 0.10 |
ENST00000557598.1
ENST00000556064.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr5_-_95158375 | 0.09 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr5_-_61031495 | 0.09 |
ENST00000506550.1
ENST00000512882.2 |
CTD-2170G1.2
|
CTD-2170G1.2 |
| chr5_-_124084493 | 0.08 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr6_-_52926539 | 0.08 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr15_+_58702742 | 0.08 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr4_-_122854193 | 0.07 |
ENST00000513531.1
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr11_+_2482661 | 0.07 |
ENST00000335475.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr5_-_179045199 | 0.07 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr11_-_57479673 | 0.07 |
ENST00000337672.2
ENST00000431606.2 |
MED19
|
mediator complex subunit 19 |
| chr6_+_42531798 | 0.07 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chrX_+_36053908 | 0.06 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr4_-_39034542 | 0.06 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr4_-_177190364 | 0.06 |
ENST00000296525.3
|
ASB5
|
ankyrin repeat and SOCS box containing 5 |
| chr7_-_107968921 | 0.06 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr4_+_81951957 | 0.06 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr14_-_92588246 | 0.06 |
ENST00000329559.3
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr14_+_50291993 | 0.06 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr7_+_120702819 | 0.05 |
ENST00000423795.1
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_+_101361107 | 0.05 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chrX_+_65384052 | 0.05 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr14_-_95922526 | 0.05 |
ENST00000554873.1
|
SYNE3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr3_+_37035289 | 0.05 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr20_+_43343886 | 0.05 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr7_-_37026108 | 0.05 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr4_+_75174204 | 0.05 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr10_-_61495760 | 0.04 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr6_-_3195981 | 0.04 |
ENST00000425384.2
ENST00000435043.2 |
RP1-40E16.9
|
RP1-40E16.9 |
| chr4_-_69083720 | 0.04 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr7_-_14942944 | 0.04 |
ENST00000403951.2
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chrX_+_52927576 | 0.04 |
ENST00000416841.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr12_+_101673872 | 0.03 |
ENST00000261637.4
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast) |
| chr18_+_29769978 | 0.03 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr16_-_15149828 | 0.03 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr4_+_75174180 | 0.03 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr12_-_89746173 | 0.02 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr8_-_19459993 | 0.02 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chrX_-_71792477 | 0.02 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr12_-_1058685 | 0.01 |
ENST00000397230.2
ENST00000542785.1 ENST00000544742.1 ENST00000536177.1 ENST00000539046.1 ENST00000541619.1 |
RAD52
|
RAD52 homolog (S. cerevisiae) |
| chr3_+_38537763 | 0.01 |
ENST00000287675.5
ENST00000358249.2 ENST00000422077.2 |
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr9_+_99690592 | 0.01 |
ENST00000354649.3
|
NUTM2G
|
NUT family member 2G |
| chr7_-_73133959 | 0.01 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr14_-_38028689 | 0.01 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr5_+_140579162 | 0.01 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr4_-_122854612 | 0.01 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr6_-_121655850 | 0.00 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr8_+_101170563 | 0.00 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr8_-_94029882 | 0.00 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr17_-_65235916 | 0.00 |
ENST00000579861.1
|
HELZ
|
helicase with zinc finger |
| chr6_-_112575758 | 0.00 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr7_+_34697973 | 0.00 |
ENST00000381539.3
|
NPSR1
|
neuropeptide S receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.2 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.3 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.0 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.7 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0097181 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0005715 | late recombination nodule(GO:0005715) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.2 | GO:0052811 | 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |