Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
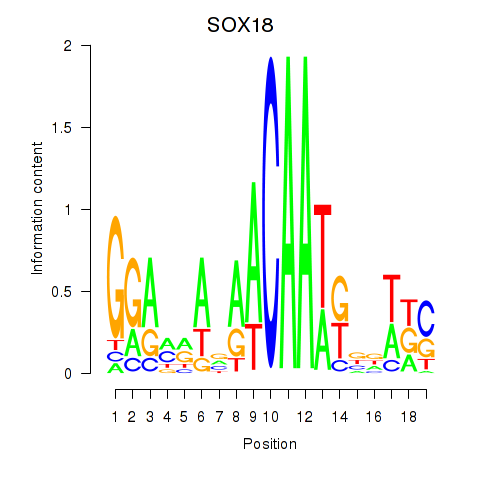
Results for SOX18
Z-value: 0.95
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | -0.57 | 4.3e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_5711010 | 1.03 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr17_+_41158742 | 0.85 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr11_+_5710919 | 0.85 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr11_-_104817919 | 0.68 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_24126350 | 0.53 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr2_+_101591314 | 0.50 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr2_-_158345341 | 0.44 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr12_+_113344582 | 0.43 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr8_-_123793048 | 0.41 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr19_+_4402659 | 0.40 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr22_+_31488433 | 0.40 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr12_+_113344755 | 0.38 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_122283424 | 0.37 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr6_-_33754778 | 0.37 |
ENST00000508327.1
ENST00000513701.1 |
LEMD2
|
LEM domain containing 2 |
| chr17_-_54893250 | 0.36 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr1_+_55446465 | 0.36 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr12_-_27235447 | 0.35 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr3_+_188817891 | 0.35 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr22_-_31324215 | 0.35 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr19_+_36132631 | 0.35 |
ENST00000379026.2
ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2
|
ets variant 2 |
| chr1_-_155880672 | 0.35 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr5_-_136834982 | 0.35 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr1_-_11918988 | 0.31 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr12_+_69633407 | 0.27 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_+_15475462 | 0.26 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_+_75628232 | 0.26 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr16_+_48657361 | 0.26 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr5_+_32585605 | 0.26 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr3_-_146262637 | 0.25 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr16_-_68034470 | 0.25 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr17_+_72427477 | 0.24 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chrX_-_49041242 | 0.24 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr7_+_35756092 | 0.24 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr11_+_71276609 | 0.24 |
ENST00000398531.1
ENST00000376536.4 |
KRTAP5-10
|
keratin associated protein 5-10 |
| chr8_+_119294456 | 0.24 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr2_+_217277137 | 0.24 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr19_+_39971505 | 0.23 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr12_-_10562356 | 0.23 |
ENST00000309384.1
|
KLRC4
|
killer cell lectin-like receptor subfamily C, member 4 |
| chr6_+_123317116 | 0.23 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr12_+_104697504 | 0.23 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr8_+_24151620 | 0.22 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr2_-_208890218 | 0.22 |
ENST00000457206.1
ENST00000427836.2 ENST00000389247.4 |
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_-_158345462 | 0.22 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr6_-_133035185 | 0.22 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr3_+_145782358 | 0.22 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr17_-_17184605 | 0.22 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chrX_+_100645812 | 0.22 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr19_+_39971470 | 0.21 |
ENST00000607714.1
ENST00000599794.1 ENST00000597666.1 ENST00000601403.1 ENST00000602028.1 |
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr6_-_27880174 | 0.21 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr13_-_46756351 | 0.21 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr13_-_46716969 | 0.20 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_-_31125850 | 0.20 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr10_-_4285923 | 0.19 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr5_+_57878859 | 0.19 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr5_-_55412774 | 0.19 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr11_-_34379546 | 0.19 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr11_-_118966167 | 0.19 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr3_+_57882061 | 0.19 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr3_+_10312604 | 0.18 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr12_-_56321649 | 0.18 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr19_+_41256764 | 0.18 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_210517895 | 0.18 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_+_109541772 | 0.18 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr8_-_21669826 | 0.18 |
ENST00000517328.1
|
GFRA2
|
GDNF family receptor alpha 2 |
| chr19_+_33571786 | 0.18 |
ENST00000170564.2
|
GPATCH1
|
G patch domain containing 1 |
| chr10_-_73497581 | 0.18 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr2_-_114647327 | 0.17 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr3_+_19189946 | 0.17 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr1_-_201123586 | 0.17 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr2_+_210518057 | 0.17 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_-_201123546 | 0.17 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr9_+_112887772 | 0.17 |
ENST00000259318.7
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr17_-_76719638 | 0.16 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr6_-_134639180 | 0.16 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_-_127309521 | 0.16 |
ENST00000469111.1
|
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr18_+_74240610 | 0.16 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr17_+_6658878 | 0.16 |
ENST00000574394.1
|
XAF1
|
XIAP associated factor 1 |
| chr16_+_28770025 | 0.15 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr15_+_75628419 | 0.15 |
ENST00000567377.1
ENST00000562789.1 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr5_+_32711829 | 0.15 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_+_225600404 | 0.15 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr4_+_109541740 | 0.15 |
ENST00000394665.1
|
RPL34
|
ribosomal protein L34 |
| chr10_-_13390270 | 0.15 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr15_+_44084040 | 0.15 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr14_+_24458123 | 0.15 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr9_+_137987825 | 0.14 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr12_-_70093235 | 0.14 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr16_+_48657337 | 0.14 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chr17_+_12569472 | 0.14 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr15_+_28624878 | 0.14 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr6_-_75953484 | 0.14 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr5_-_146461027 | 0.14 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr15_-_55541227 | 0.13 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_41166414 | 0.13 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr17_+_41150290 | 0.13 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr17_+_41150479 | 0.13 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr7_-_122840015 | 0.13 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr6_-_26235206 | 0.13 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr7_+_141408153 | 0.13 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr11_-_85397167 | 0.13 |
ENST00000316398.3
|
CCDC89
|
coiled-coil domain containing 89 |
| chr3_-_120003941 | 0.13 |
ENST00000464295.1
|
GPR156
|
G protein-coupled receptor 156 |
| chr8_-_102218292 | 0.13 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr15_+_67418047 | 0.12 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr3_+_190281229 | 0.12 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr6_-_33385823 | 0.12 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr1_+_10459433 | 0.12 |
ENST00000465632.1
ENST00000460189.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr2_+_217277466 | 0.12 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr6_-_33385655 | 0.12 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr15_-_30261066 | 0.12 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr1_+_8021713 | 0.12 |
ENST00000338639.5
ENST00000493678.1 ENST00000377493.5 |
PARK7
|
parkinson protein 7 |
| chr19_-_39805976 | 0.12 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr1_-_45988542 | 0.12 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr7_-_148725733 | 0.12 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr22_-_22292934 | 0.11 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr5_-_39462390 | 0.11 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_47004599 | 0.11 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr5_+_140800638 | 0.11 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr15_+_75628394 | 0.11 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr17_+_6916527 | 0.11 |
ENST00000552321.1
|
RNASEK
|
ribonuclease, RNase K |
| chr7_+_99933730 | 0.11 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr3_+_57881966 | 0.11 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chrX_+_47004639 | 0.11 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr4_+_110736659 | 0.11 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr9_+_125437315 | 0.11 |
ENST00000304820.2
|
OR1L3
|
olfactory receptor, family 1, subfamily L, member 3 |
| chr12_+_110338323 | 0.11 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr4_+_26322987 | 0.11 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr3_+_57741957 | 0.10 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr18_+_3447572 | 0.10 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_+_110940111 | 0.10 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chrX_-_154033661 | 0.10 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr12_-_104443890 | 0.10 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr2_-_202562774 | 0.10 |
ENST00000396886.3
ENST00000409143.1 |
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr9_-_25678856 | 0.10 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr11_+_113258495 | 0.10 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr10_-_85985294 | 0.10 |
ENST00000538192.1
|
LRIT2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr15_-_68497657 | 0.10 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr12_+_96252706 | 0.10 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr11_-_89224299 | 0.09 |
ENST00000343727.5
ENST00000531342.1 ENST00000375979.3 |
NOX4
|
NADPH oxidase 4 |
| chr1_+_209929494 | 0.09 |
ENST00000367026.3
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr17_-_28661065 | 0.09 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr5_+_99871004 | 0.09 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr4_+_144312659 | 0.09 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr15_-_59500973 | 0.09 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr7_-_45026419 | 0.09 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr1_+_209929377 | 0.09 |
ENST00000400959.3
ENST00000367025.3 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr22_+_45714672 | 0.08 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr10_-_127505167 | 0.08 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr12_+_110940005 | 0.08 |
ENST00000409246.1
ENST00000392672.4 ENST00000409300.1 ENST00000409425.1 |
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr2_+_87135076 | 0.08 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr19_-_6501778 | 0.08 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chrX_+_36254051 | 0.08 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_+_110338063 | 0.08 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr11_-_76155700 | 0.08 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr8_+_97773457 | 0.08 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr21_-_15918618 | 0.08 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr3_+_57882024 | 0.08 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr3_+_101818088 | 0.08 |
ENST00000491959.1
|
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr2_+_74881355 | 0.08 |
ENST00000357877.2
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr15_+_91473403 | 0.08 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr2_-_100939195 | 0.08 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr11_+_26495447 | 0.08 |
ENST00000531568.1
|
ANO3
|
anoctamin 3 |
| chr8_-_37707356 | 0.08 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr6_-_33385870 | 0.07 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_+_134201911 | 0.07 |
ENST00000389881.3
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chrX_+_101470280 | 0.07 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr11_-_75236867 | 0.07 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr6_+_25652501 | 0.07 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr6_-_88032795 | 0.07 |
ENST00000296882.3
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr4_+_69962185 | 0.07 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr5_+_140864649 | 0.07 |
ENST00000306593.1
|
PCDHGC4
|
protocadherin gamma subfamily C, 4 |
| chr3_-_11762202 | 0.07 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr13_-_33780133 | 0.07 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr3_+_183814852 | 0.07 |
ENST00000415389.2
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3E, ionotropic |
| chr7_-_6048650 | 0.07 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr17_-_8661860 | 0.07 |
ENST00000328794.6
|
SPDYE4
|
speedy/RINGO cell cycle regulator family member E4 |
| chr9_-_128246769 | 0.07 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr5_+_115358088 | 0.07 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr17_+_48503603 | 0.07 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr17_+_48503519 | 0.07 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr7_-_122339162 | 0.07 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chrX_-_101726732 | 0.07 |
ENST00000457521.2
ENST00000412230.2 ENST00000453326.2 |
NXF2B
TCP11X2
|
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr1_-_153517473 | 0.06 |
ENST00000368715.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr9_-_130497565 | 0.06 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr14_-_80697396 | 0.06 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr11_+_74699942 | 0.06 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr9_-_127533519 | 0.06 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr8_+_55528627 | 0.06 |
ENST00000220676.1
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr4_+_164265035 | 0.06 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr6_-_160679905 | 0.06 |
ENST00000366953.3
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr19_-_12708113 | 0.06 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr4_+_109541722 | 0.06 |
ENST00000394667.3
ENST00000502534.1 |
RPL34
|
ribosomal protein L34 |
| chr12_+_3000073 | 0.06 |
ENST00000397132.2
|
TULP3
|
tubby like protein 3 |
| chr2_-_238305397 | 0.06 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chrM_+_8527 | 0.06 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr2_-_40739501 | 0.06 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr5_-_179050066 | 0.06 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr2_-_165811756 | 0.06 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr22_+_37678505 | 0.05 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr12_-_71148413 | 0.05 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr12_-_75603202 | 0.05 |
ENST00000393288.2
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:1903093 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 1.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 1.8 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.0 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.7 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |