Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
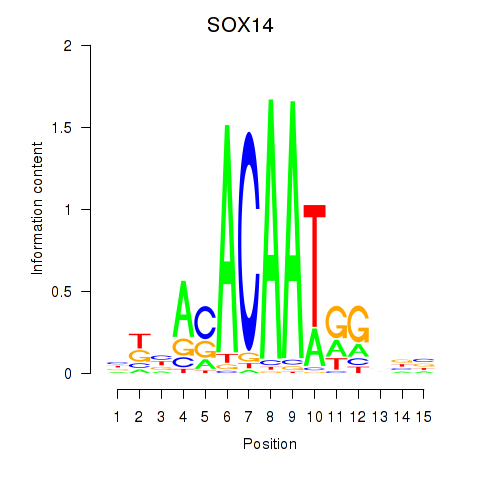
Results for SOX14
Z-value: 0.70
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Activity profile of SOX14 motif
Sorted Z-values of SOX14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_106959631 | 0.60 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr12_-_71551868 | 0.53 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr12_-_58145889 | 0.43 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr5_-_111754948 | 0.42 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr17_-_8113542 | 0.37 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr6_-_31697977 | 0.36 |
ENST00000375787.2
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_28875126 | 0.34 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr11_-_63684316 | 0.31 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr16_+_28875268 | 0.31 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr5_+_111755280 | 0.29 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr12_-_49581152 | 0.29 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_32132360 | 0.29 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr6_+_30687978 | 0.28 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr15_+_52311398 | 0.25 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr11_-_62414070 | 0.24 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr2_-_204400013 | 0.24 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_-_37382683 | 0.24 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr22_+_50247449 | 0.23 |
ENST00000216268.5
|
ZBED4
|
zinc finger, BED-type containing 4 |
| chr12_-_122907091 | 0.23 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr5_+_95066823 | 0.22 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr14_+_52313833 | 0.22 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_52521928 | 0.22 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr7_+_134551583 | 0.22 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr3_-_47484661 | 0.22 |
ENST00000495603.2
|
SCAP
|
SREBF chaperone |
| chr6_-_86353510 | 0.22 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_225616515 | 0.21 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr3_+_51851612 | 0.21 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr15_+_71184931 | 0.21 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_52521797 | 0.20 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr16_-_52580920 | 0.19 |
ENST00000219746.9
|
TOX3
|
TOX high mobility group box family member 3 |
| chr17_+_27071002 | 0.18 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr12_-_58145604 | 0.18 |
ENST00000552254.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr6_+_10556215 | 0.18 |
ENST00000316170.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr19_+_49468558 | 0.18 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr17_+_68100989 | 0.18 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr2_+_239756671 | 0.18 |
ENST00000448943.2
|
TWIST2
|
twist family bHLH transcription factor 2 |
| chrX_-_15872914 | 0.18 |
ENST00000380291.1
ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_-_62308087 | 0.18 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr3_+_164924716 | 0.17 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr17_+_27920486 | 0.17 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr1_-_26232522 | 0.17 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr1_+_226250379 | 0.17 |
ENST00000366815.3
ENST00000366814.3 |
H3F3A
|
H3 histone, family 3A |
| chrX_-_38080077 | 0.17 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr11_+_4116005 | 0.17 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr7_+_11013491 | 0.16 |
ENST00000403050.3
ENST00000445996.2 |
PHF14
|
PHD finger protein 14 |
| chr3_+_187957646 | 0.16 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_130184470 | 0.16 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr1_+_52521957 | 0.16 |
ENST00000472944.2
ENST00000484036.1 |
BTF3L4
|
basic transcription factor 3-like 4 |
| chr1_-_26232951 | 0.16 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr18_+_42260861 | 0.16 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr1_-_52521831 | 0.15 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr14_-_100772767 | 0.15 |
ENST00000392908.3
ENST00000539621.1 |
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr11_+_4116054 | 0.15 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr2_-_157198860 | 0.15 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr2_-_204398141 | 0.15 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr17_-_53046058 | 0.15 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr3_-_129513259 | 0.15 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr11_-_44971702 | 0.14 |
ENST00000533940.1
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr17_-_8113886 | 0.14 |
ENST00000577833.1
ENST00000534871.1 ENST00000583915.1 ENST00000316199.6 ENST00000581511.1 ENST00000585124.1 |
AURKB
|
aurora kinase B |
| chr2_+_240323439 | 0.14 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr16_+_28874860 | 0.14 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr6_+_122720681 | 0.13 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr15_+_73976545 | 0.13 |
ENST00000318443.5
ENST00000537340.2 ENST00000318424.5 |
CD276
|
CD276 molecule |
| chr1_+_93913713 | 0.13 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr2_-_9770706 | 0.13 |
ENST00000381844.4
|
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr10_+_123923205 | 0.13 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr22_-_19512893 | 0.13 |
ENST00000403084.1
ENST00000413119.2 |
CLDN5
|
claudin 5 |
| chr1_-_204463829 | 0.13 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr9_+_128509624 | 0.13 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr3_+_49840685 | 0.13 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr14_+_74815116 | 0.13 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr2_+_65215604 | 0.12 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr14_+_76776957 | 0.12 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr12_-_15942503 | 0.12 |
ENST00000281172.5
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr14_-_21567009 | 0.12 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr10_-_13390270 | 0.12 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr14_-_100772796 | 0.12 |
ENST00000554060.1
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr14_-_100772862 | 0.12 |
ENST00000359232.3
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr1_-_204436344 | 0.12 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr1_-_161014731 | 0.12 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr1_+_33231268 | 0.12 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr10_+_123922941 | 0.12 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr14_+_96829886 | 0.12 |
ENST00000556095.1
|
GSKIP
|
GSK3B interacting protein |
| chr5_+_140514782 | 0.12 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr6_+_170190417 | 0.11 |
ENST00000420557.2
|
LINC00574
|
long intergenic non-protein coding RNA 574 |
| chr12_+_69633372 | 0.11 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr7_+_30068260 | 0.11 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr11_+_66247880 | 0.11 |
ENST00000360510.2
ENST00000453114.1 ENST00000541961.1 ENST00000532019.1 ENST00000526515.1 ENST00000530165.1 ENST00000533725.1 |
DPP3
|
dipeptidyl-peptidase 3 |
| chr10_+_18429606 | 0.11 |
ENST00000324631.7
ENST00000352115.6 ENST00000377328.1 |
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_-_35492048 | 0.11 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr2_-_160472052 | 0.10 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr10_+_123923105 | 0.10 |
ENST00000368999.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_30848557 | 0.10 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr2_-_204399976 | 0.10 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr21_+_35552978 | 0.10 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr19_-_58514129 | 0.10 |
ENST00000552184.1
ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606
|
zinc finger protein 606 |
| chr10_+_89622870 | 0.10 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr17_-_48207157 | 0.10 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr3_+_186649133 | 0.10 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_+_186358148 | 0.10 |
ENST00000382134.3
ENST00000265029.3 |
FETUB
|
fetuin B |
| chr18_+_74240756 | 0.10 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr12_-_15942309 | 0.09 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr13_+_111767650 | 0.09 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_-_2418596 | 0.09 |
ENST00000381218.3
|
ZBED1
|
zinc finger, BED-type containing 1 |
| chrX_-_2418634 | 0.09 |
ENST00000444280.1
|
DHRSX
|
dehydrogenase/reductase (SDR family) X-linked |
| chr14_+_21566980 | 0.09 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr17_-_42100474 | 0.09 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr5_+_66124590 | 0.09 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_65996599 | 0.09 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr3_-_57583185 | 0.09 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr19_+_58514229 | 0.09 |
ENST00000546949.1
ENST00000553254.1 ENST00000547364.1 |
CTD-2368P22.1
|
HCG1811579; Uncharacterized protein |
| chr10_+_69644404 | 0.09 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr11_-_62477041 | 0.09 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr6_+_32937083 | 0.08 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr21_-_35340759 | 0.08 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr12_-_105478339 | 0.08 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr10_-_21806759 | 0.08 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr14_-_96830207 | 0.08 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr7_+_30067973 | 0.08 |
ENST00000258679.7
ENST00000449726.1 ENST00000396257.2 ENST00000396259.1 |
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr17_-_14683517 | 0.08 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr9_-_100954910 | 0.08 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr5_-_134734901 | 0.08 |
ENST00000312469.4
ENST00000423969.2 |
H2AFY
|
H2A histone family, member Y |
| chr13_-_45915221 | 0.08 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr2_+_54350316 | 0.08 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_9771075 | 0.08 |
ENST00000446619.1
ENST00000238081.3 |
YWHAQ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr7_-_149470297 | 0.07 |
ENST00000484747.1
|
ZNF467
|
zinc finger protein 467 |
| chr8_+_133787586 | 0.07 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr19_+_38397839 | 0.07 |
ENST00000222345.6
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr14_+_96829814 | 0.07 |
ENST00000555181.1
ENST00000553699.1 ENST00000554182.1 |
GSKIP
|
GSK3B interacting protein |
| chr17_+_68101117 | 0.07 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_-_130184555 | 0.07 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr8_+_144295067 | 0.07 |
ENST00000330824.2
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr20_-_44991813 | 0.07 |
ENST00000372227.1
|
SLC35C2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr8_-_110620284 | 0.07 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr3_-_101232019 | 0.07 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr11_-_36310958 | 0.07 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr11_+_10772534 | 0.07 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr15_+_73976715 | 0.07 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr1_-_201123586 | 0.07 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr1_-_152196669 | 0.07 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr16_+_30406721 | 0.07 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr12_-_58146128 | 0.07 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr11_+_111807863 | 0.07 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr20_+_44036620 | 0.07 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr17_+_15603447 | 0.07 |
ENST00000395893.2
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr10_+_70480963 | 0.07 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr9_-_130712995 | 0.06 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr22_+_23522552 | 0.06 |
ENST00000359540.3
ENST00000398512.5 |
BCR
|
breakpoint cluster region |
| chr16_+_57680043 | 0.06 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_40915725 | 0.06 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr1_-_40782938 | 0.06 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr10_-_32217717 | 0.06 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr19_+_7580944 | 0.06 |
ENST00000597229.1
|
ZNF358
|
zinc finger protein 358 |
| chr12_-_498620 | 0.06 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr12_-_58146048 | 0.06 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr6_-_3227877 | 0.06 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr17_+_1666108 | 0.05 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_48673552 | 0.05 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chr20_+_58179582 | 0.05 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr6_+_114178512 | 0.05 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr17_+_67498396 | 0.05 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_201123546 | 0.05 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr3_-_182698381 | 0.05 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr12_+_498500 | 0.05 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr15_+_64388166 | 0.05 |
ENST00000353874.4
ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1
|
sorting nexin 1 |
| chr11_+_19798964 | 0.05 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr18_+_74240610 | 0.05 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr15_-_86338100 | 0.05 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr4_+_150999418 | 0.05 |
ENST00000296550.7
|
DCLK2
|
doublecortin-like kinase 2 |
| chr5_-_179050066 | 0.05 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_+_64238539 | 0.05 |
ENST00000357825.3
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr17_-_39041479 | 0.05 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chrX_+_100646190 | 0.05 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr4_-_120548779 | 0.04 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr2_-_55276320 | 0.04 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr4_+_146403912 | 0.04 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr3_+_186358200 | 0.04 |
ENST00000382136.3
|
FETUB
|
fetuin B |
| chr20_+_44036900 | 0.04 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr8_+_24151553 | 0.04 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr15_-_58571445 | 0.04 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr22_+_29664241 | 0.04 |
ENST00000436425.1
ENST00000447973.1 |
EWSR1
|
EWS RNA-binding protein 1 |
| chr2_-_217560248 | 0.04 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr12_+_99038919 | 0.04 |
ENST00000551964.1
|
APAF1
|
apoptotic peptidase activating factor 1 |
| chr9_-_116062045 | 0.04 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183 |
| chr7_+_69064300 | 0.04 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr16_-_67217844 | 0.04 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr14_-_21994525 | 0.04 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr19_+_10765699 | 0.04 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chrX_+_70503433 | 0.04 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr11_-_62476965 | 0.04 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr8_-_134501937 | 0.04 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr12_+_69004619 | 0.04 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr1_+_14075865 | 0.04 |
ENST00000413440.1
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr17_-_67138015 | 0.03 |
ENST00000284425.2
ENST00000590645.1 |
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr22_+_38054721 | 0.03 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr18_+_20715416 | 0.03 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr5_-_137071756 | 0.03 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr16_-_49890016 | 0.03 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr12_+_69633407 | 0.03 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_+_98909260 | 0.03 |
ENST00000556029.1
|
TMPO
|
thymopoietin |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990575 | mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.5 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.7 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.2 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.2 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:1990619 | histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.2 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:1904879 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0034371 | positive regulation of lipoprotein particle clearance(GO:0010986) chylomicron remodeling(GO:0034371) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0015186 | L-cystine transmembrane transporter activity(GO:0015184) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |