Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
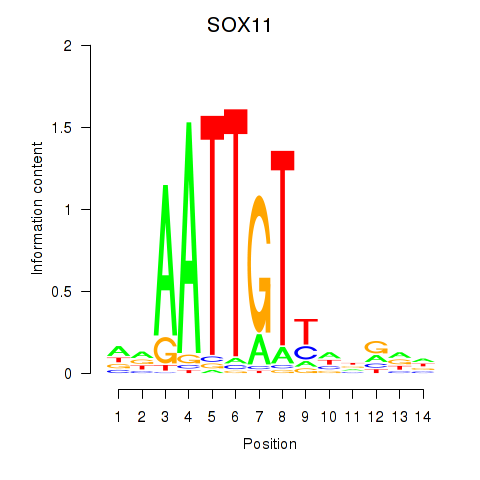
Results for SOX11
Z-value: 0.46
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX11 | hg19_v2_chr2_+_5832799_5832799 | -0.43 | 5.7e-01 | Click! |
Activity profile of SOX11 motif
Sorted Z-values of SOX11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_71403280 | 0.25 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr2_-_37068530 | 0.24 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr6_+_26183958 | 0.22 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr8_-_25281747 | 0.21 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_+_207804278 | 0.20 |
ENST00000272852.3
|
CPO
|
carboxypeptidase O |
| chr11_-_58611957 | 0.17 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr20_-_7921090 | 0.17 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr21_+_17792672 | 0.15 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_105416039 | 0.15 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr15_+_80733570 | 0.15 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr11_-_58612168 | 0.14 |
ENST00000287275.1
|
GLYATL2
|
glycine-N-acyltransferase-like 2 |
| chr8_+_67104323 | 0.14 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr4_+_110749143 | 0.14 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr16_+_67381263 | 0.13 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chrX_-_132231123 | 0.13 |
ENST00000511190.1
|
USP26
|
ubiquitin specific peptidase 26 |
| chr2_+_166326157 | 0.12 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chrX_+_100645812 | 0.12 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr7_+_66800928 | 0.12 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr4_+_57276661 | 0.12 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr13_+_78315466 | 0.12 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr13_+_78315348 | 0.11 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_-_30585009 | 0.11 |
ENST00000376511.2
|
PPP1R10
|
protein phosphatase 1, regulatory subunit 10 |
| chr7_-_143579973 | 0.11 |
ENST00000460532.1
ENST00000491908.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr6_-_11779014 | 0.11 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chrX_-_13835147 | 0.11 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr6_-_31125850 | 0.11 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr3_+_112930387 | 0.10 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr5_+_172571445 | 0.10 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr17_-_72772425 | 0.10 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr5_+_81601166 | 0.10 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr5_+_179159813 | 0.10 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr10_-_25305011 | 0.10 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr2_+_42104692 | 0.10 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr4_+_71019903 | 0.10 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr13_-_45048386 | 0.10 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr6_-_11779174 | 0.10 |
ENST00000379413.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr4_-_71532207 | 0.09 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_-_71532339 | 0.09 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_-_38036271 | 0.09 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr11_+_47293795 | 0.09 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr19_+_50094866 | 0.09 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr3_+_113251143 | 0.09 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr18_+_22040620 | 0.09 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr19_+_39989535 | 0.09 |
ENST00000356433.5
|
DLL3
|
delta-like 3 (Drosophila) |
| chr11_+_57425209 | 0.09 |
ENST00000533905.1
ENST00000525602.1 ENST00000302731.4 |
CLP1
|
cleavage and polyadenylation factor I subunit 1 |
| chr15_+_69857515 | 0.08 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr19_-_6501778 | 0.08 |
ENST00000596291.1
|
TUBB4A
|
tubulin, beta 4A class IVa |
| chrX_+_23928500 | 0.08 |
ENST00000435707.1
|
CXorf58
|
chromosome X open reading frame 58 |
| chr7_+_98923505 | 0.08 |
ENST00000432884.2
ENST00000262942.5 |
ARPC1A
|
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr6_-_49712091 | 0.08 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_-_47950745 | 0.08 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr19_-_5680499 | 0.08 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr9_+_71986182 | 0.08 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr17_-_72772462 | 0.07 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr13_+_78315528 | 0.07 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_-_133055896 | 0.07 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr22_-_30642728 | 0.07 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr4_-_73434498 | 0.07 |
ENST00000286657.4
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr2_-_176046391 | 0.06 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr6_+_53883708 | 0.06 |
ENST00000514921.1
ENST00000274897.5 ENST00000370877.2 |
MLIP
|
muscular LMNA-interacting protein |
| chr15_-_64673665 | 0.06 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr12_-_22063787 | 0.06 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr8_+_41386725 | 0.06 |
ENST00000276533.3
ENST00000520710.1 ENST00000518671.1 |
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr3_+_112930373 | 0.06 |
ENST00000498710.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr16_+_67261008 | 0.06 |
ENST00000304800.9
ENST00000563953.1 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr17_+_6347729 | 0.06 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr1_-_224624730 | 0.05 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr4_-_71532601 | 0.05 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_-_24469602 | 0.05 |
ENST00000270800.1
|
IL22RA1
|
interleukin 22 receptor, alpha 1 |
| chr20_-_32308028 | 0.05 |
ENST00000409299.3
ENST00000217398.3 ENST00000344022.3 |
PXMP4
|
peroxisomal membrane protein 4, 24kDa |
| chr15_-_64673630 | 0.05 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr22_+_24407776 | 0.05 |
ENST00000405822.2
|
CABIN1
|
calcineurin binding protein 1 |
| chr12_+_41136144 | 0.05 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr21_-_45671014 | 0.05 |
ENST00000436357.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr2_+_33683109 | 0.05 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_-_115872142 | 0.05 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr10_-_127505167 | 0.05 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr8_+_41386761 | 0.04 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr1_+_149754227 | 0.04 |
ENST00000444948.1
ENST00000369168.4 |
FCGR1A
|
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr8_-_37707356 | 0.04 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr3_-_112564797 | 0.04 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr6_+_53883790 | 0.04 |
ENST00000509997.1
|
MLIP
|
muscular LMNA-interacting protein |
| chr3_-_18466026 | 0.04 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr3_+_113616317 | 0.04 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr7_-_56118981 | 0.04 |
ENST00000419984.2
ENST00000413218.1 ENST00000424596.1 |
PSPH
|
phosphoserine phosphatase |
| chr7_-_7575477 | 0.04 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr15_-_79383102 | 0.04 |
ENST00000558480.2
ENST00000419573.3 |
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr10_+_69865866 | 0.04 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr17_-_79895097 | 0.04 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr9_-_95166841 | 0.04 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr6_-_36515177 | 0.04 |
ENST00000229812.7
|
STK38
|
serine/threonine kinase 38 |
| chr2_+_138721850 | 0.04 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr6_-_32557610 | 0.04 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr2_+_102928009 | 0.04 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr1_+_146373546 | 0.04 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr11_+_12399071 | 0.04 |
ENST00000539723.1
ENST00000550549.1 |
PARVA
|
parvin, alpha |
| chr11_+_120973375 | 0.03 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr21_+_43619796 | 0.03 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr16_-_28192360 | 0.03 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr17_-_66951382 | 0.03 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr17_-_47786375 | 0.03 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr10_+_35456444 | 0.03 |
ENST00000361599.4
|
CREM
|
cAMP responsive element modulator |
| chr3_+_130279178 | 0.03 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr8_+_24241789 | 0.03 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr11_+_61976137 | 0.03 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr17_-_79895154 | 0.03 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr9_+_12775011 | 0.03 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr15_+_49447947 | 0.03 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr10_-_95241951 | 0.03 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr4_+_169013666 | 0.03 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr6_-_76072719 | 0.03 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr17_-_66951474 | 0.03 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr5_+_140753444 | 0.03 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr5_+_140248518 | 0.03 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr7_-_81399355 | 0.03 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_-_32173579 | 0.03 |
ENST00000359836.1
ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD
|
dystrophin |
| chr15_-_78913628 | 0.02 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr7_-_81399329 | 0.02 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_-_15619076 | 0.02 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr6_-_49712147 | 0.02 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr5_-_86534822 | 0.02 |
ENST00000445770.2
|
AC008394.1
|
Uncharacterized protein |
| chr12_-_11287243 | 0.02 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr7_+_18536090 | 0.02 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr1_+_207038699 | 0.02 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr18_+_46065393 | 0.02 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr10_-_95242044 | 0.02 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr6_-_49712072 | 0.02 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr9_-_21142144 | 0.02 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr12_+_56075330 | 0.02 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr3_+_112930306 | 0.02 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr4_+_144312659 | 0.02 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_+_145524891 | 0.02 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr20_+_35807512 | 0.02 |
ENST00000373622.5
|
RPN2
|
ribophorin II |
| chr11_+_22688150 | 0.02 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chrM_+_8527 | 0.02 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr8_+_24241969 | 0.02 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chrX_+_107288197 | 0.02 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_-_170893268 | 0.02 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2 |
| chr20_-_5426380 | 0.02 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr18_-_67614645 | 0.02 |
ENST00000577287.1
|
CD226
|
CD226 molecule |
| chrX_+_107288239 | 0.02 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr11_+_92085262 | 0.02 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr6_-_32977345 | 0.02 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr5_+_140207536 | 0.02 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chrX_+_27826107 | 0.02 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chrX_-_73513307 | 0.02 |
ENST00000602420.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr12_+_32655048 | 0.02 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr17_+_62461569 | 0.02 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chr2_-_219157250 | 0.01 |
ENST00000434015.2
ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr7_+_90032821 | 0.01 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr11_-_64684672 | 0.01 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr1_+_204839959 | 0.01 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr7_-_81399287 | 0.01 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr5_+_148521454 | 0.01 |
ENST00000508983.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr15_+_26147507 | 0.01 |
ENST00000383019.2
ENST00000557171.1 |
RP11-1084I9.1
|
RP11-1084I9.1 |
| chr3_-_151160938 | 0.01 |
ENST00000489791.1
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr2_-_71222466 | 0.01 |
ENST00000606025.1
|
AC007040.11
|
Uncharacterized protein |
| chr7_-_82792215 | 0.01 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr17_+_6347761 | 0.01 |
ENST00000250056.8
ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A
|
family with sequence similarity 64, member A |
| chr10_+_32873190 | 0.01 |
ENST00000375025.4
|
C10orf68
|
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr2_-_145275828 | 0.01 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr13_+_78315295 | 0.01 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr7_-_81399411 | 0.01 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_+_30585486 | 0.01 |
ENST00000259873.4
ENST00000506373.2 |
MRPS18B
|
mitochondrial ribosomal protein S18B |
| chr20_-_30539773 | 0.01 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr1_+_145525015 | 0.01 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr17_-_10017864 | 0.01 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr1_+_149239529 | 0.01 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chrX_-_100548045 | 0.01 |
ENST00000372907.3
ENST00000372905.2 |
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chrX_+_102192200 | 0.00 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr16_+_67381289 | 0.00 |
ENST00000435835.3
|
LRRC36
|
leucine rich repeat containing 36 |
| chr1_+_214776516 | 0.00 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr18_+_7754957 | 0.00 |
ENST00000400053.4
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr9_-_114521783 | 0.00 |
ENST00000394779.3
ENST00000394777.4 |
C9orf84
|
chromosome 9 open reading frame 84 |
| chr12_-_772901 | 0.00 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr8_-_13134045 | 0.00 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr8_-_54752406 | 0.00 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr8_+_104831472 | 0.00 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr1_-_241799232 | 0.00 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr4_+_159727222 | 0.00 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr8_-_128960591 | 0.00 |
ENST00000539634.1
|
TMEM75
|
transmembrane protein 75 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX11
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.1 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.3 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |