Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for SIX6
Z-value: 1.11
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX homeobox 6 |
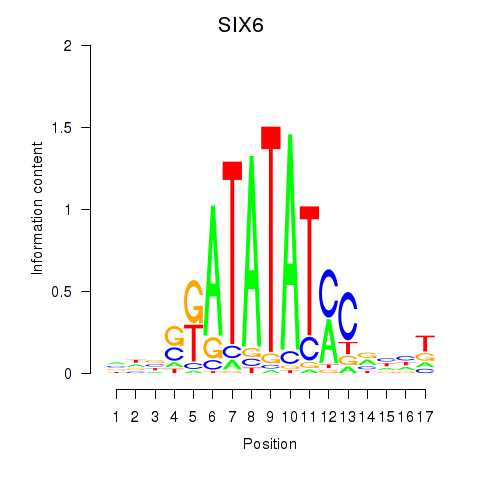
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_91061712 | 2.89 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr12_-_121477039 | 2.69 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr12_-_121476959 | 2.49 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr6_+_71104588 | 1.77 |
ENST00000418403.1
|
RP11-462G2.1
|
RP11-462G2.1 |
| chr11_-_104916034 | 1.50 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr12_-_121476750 | 1.27 |
ENST00000543677.1
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr8_-_128231299 | 0.97 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr15_-_80263506 | 0.94 |
ENST00000335661.6
|
BCL2A1
|
BCL2-related protein A1 |
| chr11_-_104972158 | 0.86 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_+_37012607 | 0.81 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr5_-_78281775 | 0.73 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr22_+_24666763 | 0.72 |
ENST00000437398.1
ENST00000421374.1 ENST00000314328.9 ENST00000541492.1 |
SPECC1L
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr4_-_71532601 | 0.62 |
ENST00000510614.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_+_189321881 | 0.56 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr19_-_53426700 | 0.56 |
ENST00000596623.1
|
ZNF888
|
zinc finger protein 888 |
| chr6_-_26043885 | 0.52 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr4_-_184241927 | 0.51 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr4_+_74301880 | 0.48 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr1_-_65468148 | 0.42 |
ENST00000415842.1
|
RP11-182I10.3
|
RP11-182I10.3 |
| chr14_-_46185155 | 0.40 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr21_-_43735628 | 0.35 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr5_-_177207634 | 0.35 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr2_-_100925967 | 0.35 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr8_-_124553437 | 0.34 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr5_+_175490540 | 0.32 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr2_+_62132800 | 0.32 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr1_-_161015752 | 0.32 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr8_+_1993152 | 0.31 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr14_+_20937538 | 0.29 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chrX_-_77225135 | 0.29 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr17_+_61271355 | 0.29 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_+_62132781 | 0.27 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr9_-_67786624 | 0.27 |
ENST00000455764.2
|
FAM27E3
|
family with sequence similarity 27, member E3 |
| chr2_-_228497888 | 0.27 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr11_+_74699703 | 0.26 |
ENST00000529024.1
ENST00000544263.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr2_+_85822857 | 0.25 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr8_+_1993173 | 0.25 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr12_+_40787194 | 0.25 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr1_-_157014865 | 0.25 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_+_151531859 | 0.25 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr2_-_239140011 | 0.24 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr11_+_122709200 | 0.24 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr16_+_28648975 | 0.24 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr16_-_29517141 | 0.24 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr12_+_119616447 | 0.24 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr6_+_127898312 | 0.23 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr17_+_60501228 | 0.23 |
ENST00000311506.5
|
METTL2A
|
methyltransferase like 2A |
| chr19_-_44031375 | 0.23 |
ENST00000292147.2
|
ETHE1
|
ethylmalonic encephalopathy 1 |
| chr4_-_71532668 | 0.23 |
ENST00000510437.1
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr4_+_74347400 | 0.23 |
ENST00000226355.3
|
AFM
|
afamin |
| chr2_+_210636697 | 0.22 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr18_+_50278430 | 0.22 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr18_+_55018044 | 0.21 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_-_150669604 | 0.21 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr1_+_160709055 | 0.21 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr9_-_100684845 | 0.21 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr10_-_79397479 | 0.20 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_259218 | 0.20 |
ENST00000449294.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr10_-_61720640 | 0.19 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chr16_-_21436459 | 0.19 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr16_+_22524844 | 0.19 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_-_21868978 | 0.19 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr22_-_17302589 | 0.18 |
ENST00000331428.5
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3 |
| chr8_+_67104323 | 0.18 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr3_-_111314230 | 0.17 |
ENST00000317012.4
|
ZBED2
|
zinc finger, BED-type containing 2 |
| chr16_-_87350970 | 0.16 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chrX_-_55020511 | 0.16 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_+_58430567 | 0.16 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr5_+_55147205 | 0.15 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr16_-_21868739 | 0.15 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr12_-_14849470 | 0.15 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr9_+_93759317 | 0.14 |
ENST00000563268.1
|
RP11-367F23.2
|
RP11-367F23.2 |
| chr16_+_21689835 | 0.14 |
ENST00000286149.4
ENST00000388958.3 |
OTOA
|
otoancorin |
| chr1_-_159684371 | 0.14 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr12_+_117348742 | 0.14 |
ENST00000309909.5
ENST00000455858.2 |
FBXW8
|
F-box and WD repeat domain containing 8 |
| chr8_+_26247878 | 0.14 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr2_+_181988560 | 0.13 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr1_+_160709076 | 0.13 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr9_-_23821842 | 0.13 |
ENST00000544538.1
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr1_+_31885963 | 0.13 |
ENST00000373709.3
|
SERINC2
|
serine incorporator 2 |
| chr7_-_141957847 | 0.12 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chr2_-_100939195 | 0.11 |
ENST00000393437.3
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr8_+_12809531 | 0.11 |
ENST00000532376.2
|
KIAA1456
|
KIAA1456 |
| chr9_-_135230336 | 0.11 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr7_+_107332334 | 0.11 |
ENST00000541474.1
ENST00000544569.1 |
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr9_-_7800067 | 0.10 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr8_+_24241789 | 0.10 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr3_-_107596910 | 0.10 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr2_-_238305397 | 0.10 |
ENST00000409809.1
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr20_+_54987305 | 0.10 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr8_+_67405438 | 0.09 |
ENST00000305454.3
|
C8orf46
|
chromosome 8 open reading frame 46 |
| chr15_+_58430368 | 0.08 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr1_+_112016414 | 0.08 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_+_63063152 | 0.08 |
ENST00000371129.3
|
ANGPTL3
|
angiopoietin-like 3 |
| chr8_-_110660975 | 0.08 |
ENST00000528045.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr22_+_18043133 | 0.08 |
ENST00000327451.6
ENST00000399813.1 |
SLC25A18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr17_-_42327236 | 0.08 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr12_-_78934441 | 0.07 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr2_-_77749387 | 0.07 |
ENST00000409884.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr8_+_24241969 | 0.07 |
ENST00000522298.1
|
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr3_+_141457030 | 0.07 |
ENST00000273480.3
|
RNF7
|
ring finger protein 7 |
| chr5_+_40909354 | 0.07 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr1_+_160709029 | 0.07 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr8_+_32579341 | 0.06 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr4_-_13546632 | 0.06 |
ENST00000382438.5
|
NKX3-2
|
NK3 homeobox 2 |
| chr2_-_77749336 | 0.06 |
ENST00000409282.1
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr11_+_124789146 | 0.06 |
ENST00000408930.5
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1 |
| chr12_-_498620 | 0.06 |
ENST00000399788.2
ENST00000382815.4 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr2_-_77749474 | 0.05 |
ENST00000409093.1
ENST00000409088.3 |
LRRTM4
|
leucine rich repeat transmembrane neuronal 4 |
| chr10_+_45495898 | 0.05 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chrX_-_53350522 | 0.05 |
ENST00000396435.3
ENST00000375368.5 |
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr21_-_19858196 | 0.04 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr11_+_126262027 | 0.04 |
ENST00000526311.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr13_+_42712178 | 0.04 |
ENST00000536612.1
|
DGKH
|
diacylglycerol kinase, eta |
| chr20_+_54987168 | 0.03 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr17_-_67057114 | 0.03 |
ENST00000370732.2
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr19_-_14628645 | 0.03 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr18_+_74240610 | 0.02 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr3_+_151531810 | 0.02 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr22_-_17702729 | 0.02 |
ENST00000449907.2
ENST00000441548.1 ENST00000399839.1 |
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr19_+_3136115 | 0.01 |
ENST00000262958.3
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chrX_+_128674213 | 0.01 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chrX_-_18238999 | 0.01 |
ENST00000380033.4
ENST00000380030.3 |
BEND2
|
BEN domain containing 2 |
| chr1_+_156119466 | 0.01 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr3_-_196242233 | 0.01 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr16_+_72090053 | 0.01 |
ENST00000576168.2
ENST00000567185.3 ENST00000567612.2 |
HP
|
haptoglobin |
| chr5_+_140201183 | 0.00 |
ENST00000529619.1
ENST00000529859.1 ENST00000378126.3 |
PCDHA5
|
protocadherin alpha 5 |
| chr1_+_40942887 | 0.00 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chrY_+_16168097 | 0.00 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 0.7 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.2 | 2.9 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.3 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.2 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 6.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.2 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 0.7 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.2 | 2.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.3 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.2 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.2 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 9.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |