Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
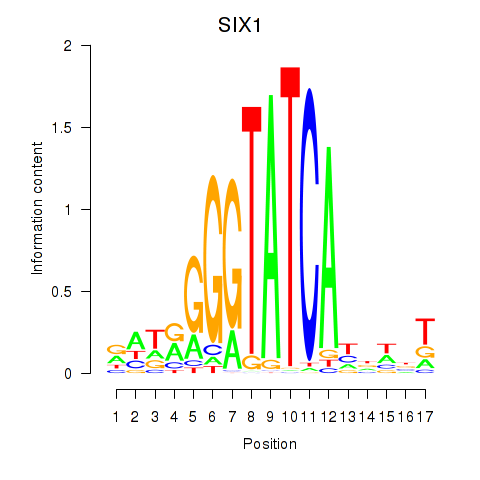
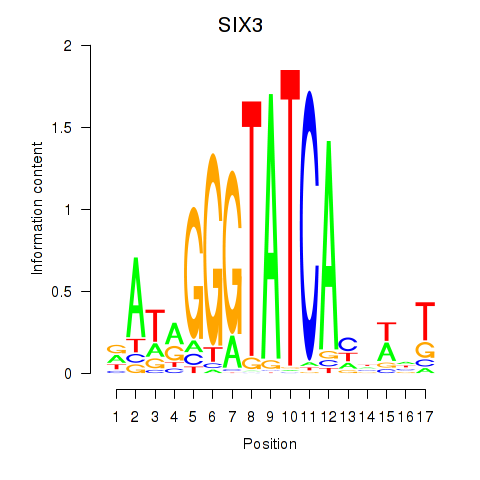
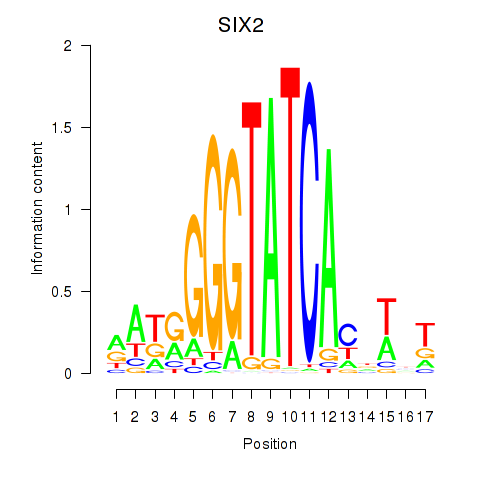
Results for SIX1_SIX3_SIX2
Z-value: 1.07
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | -0.74 | 2.6e-01 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | -0.18 | 8.2e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_241695424 | 1.40 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_-_33405600 | 1.37 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr1_+_241695670 | 1.33 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_-_31521567 | 1.10 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr13_-_46679144 | 0.97 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr5_-_159846066 | 0.84 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr13_-_46679185 | 0.81 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr16_-_28550320 | 0.76 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr7_+_139025875 | 0.65 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr6_-_136847099 | 0.64 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr6_-_136847610 | 0.63 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr17_+_4675175 | 0.63 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr16_+_22517166 | 0.62 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr9_-_34662651 | 0.61 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr16_-_28550348 | 0.60 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr15_+_75491203 | 0.59 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr3_+_149191723 | 0.57 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr4_-_140477928 | 0.57 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr9_+_67977438 | 0.55 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr2_-_150444300 | 0.54 |
ENST00000303319.5
|
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr19_+_45445524 | 0.51 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr10_-_134331695 | 0.49 |
ENST00000455414.1
|
RP11-432J24.5
|
RP11-432J24.5 |
| chr12_+_147052 | 0.48 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr1_+_64014588 | 0.46 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chr3_+_149192475 | 0.46 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_+_207277632 | 0.44 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr4_+_89300158 | 0.43 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr16_+_19098178 | 0.43 |
ENST00000568032.1
|
RP11-626G11.4
|
RP11-626G11.4 |
| chr4_-_140477910 | 0.42 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_+_214776516 | 0.40 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr12_-_10826612 | 0.39 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr15_+_59279851 | 0.39 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr1_-_109618566 | 0.38 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr1_-_207119738 | 0.38 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr8_-_95229531 | 0.36 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr17_-_7080883 | 0.36 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_-_149095652 | 0.36 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_-_467892 | 0.35 |
ENST00000506646.1
ENST00000505900.1 |
ZNF721
|
zinc finger protein 721 |
| chr11_-_8892900 | 0.34 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr14_-_74296806 | 0.34 |
ENST00000555539.1
|
RP5-1021I20.2
|
RP5-1021I20.2 |
| chr4_+_155484155 | 0.33 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_-_48068643 | 0.33 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr2_-_150444116 | 0.33 |
ENST00000428879.1
ENST00000422782.2 |
MMADHC
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr6_-_138428613 | 0.33 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr16_+_29690358 | 0.31 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr10_-_38265517 | 0.31 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chr20_-_55100981 | 0.30 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr4_+_2794750 | 0.29 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr4_+_159131346 | 0.29 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr1_+_53793885 | 0.28 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr2_+_161993465 | 0.28 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr14_+_54863667 | 0.28 |
ENST00000335183.6
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr1_+_150245099 | 0.28 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr14_+_54863682 | 0.27 |
ENST00000543789.2
ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr1_+_93913665 | 0.27 |
ENST00000271234.7
ENST00000370256.4 ENST00000260506.8 |
FNBP1L
|
formin binding protein 1-like |
| chr11_+_327171 | 0.27 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr12_-_112614506 | 0.27 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr2_+_183989157 | 0.27 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr3_+_11267691 | 0.26 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr10_-_103603523 | 0.26 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_-_53594227 | 0.26 |
ENST00000550743.2
|
ITGB7
|
integrin, beta 7 |
| chr3_+_184529929 | 0.25 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr8_+_19536083 | 0.25 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr11_-_2924720 | 0.24 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr3_+_172034218 | 0.24 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr14_+_58711539 | 0.24 |
ENST00000216455.4
ENST00000412908.2 ENST00000557508.1 |
PSMA3
|
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr11_+_85339623 | 0.23 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr11_+_17298522 | 0.23 |
ENST00000529313.1
|
NUCB2
|
nucleobindin 2 |
| chr11_+_71900572 | 0.23 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr4_-_14889791 | 0.23 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr19_+_11750566 | 0.23 |
ENST00000344893.3
|
ZNF833P
|
zinc finger protein 833, pseudogene |
| chr1_-_44818599 | 0.22 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr1_-_169337176 | 0.22 |
ENST00000472647.1
ENST00000367811.3 |
NME7
|
NME/NM23 family member 7 |
| chr19_+_11457232 | 0.22 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr9_-_113018746 | 0.22 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr6_+_96969672 | 0.22 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr22_+_21321531 | 0.21 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr1_+_150245177 | 0.21 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr14_+_91709103 | 0.20 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr15_-_100258029 | 0.20 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr2_-_105953912 | 0.20 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr12_+_16500037 | 0.20 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_128615681 | 0.20 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr14_-_94759361 | 0.20 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr1_+_215740709 | 0.20 |
ENST00000259154.4
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr2_-_220034745 | 0.20 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chrX_+_75392771 | 0.20 |
ENST00000373358.3
ENST00000373357.3 |
PBDC1
|
polysaccharide biosynthesis domain containing 1 |
| chr7_-_141541221 | 0.20 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr2_-_107502456 | 0.20 |
ENST00000419159.2
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr7_+_90012986 | 0.19 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr11_-_118134997 | 0.19 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr18_+_61575200 | 0.19 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr19_+_45445491 | 0.19 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr3_+_184529948 | 0.19 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr2_+_211421262 | 0.19 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr14_+_54863739 | 0.18 |
ENST00000541304.1
|
CDKN3
|
cyclin-dependent kinase inhibitor 3 |
| chr11_+_102552041 | 0.18 |
ENST00000537079.1
|
RP11-817J15.3
|
Uncharacterized protein |
| chr5_-_145562147 | 0.18 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr4_-_156298028 | 0.18 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr14_-_24729251 | 0.18 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr7_-_92219337 | 0.18 |
ENST00000456502.1
ENST00000427372.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr15_+_40331456 | 0.18 |
ENST00000504245.1
ENST00000560341.1 |
SRP14-AS1
|
SRP14 antisense RNA1 (head to head) |
| chr8_-_124749609 | 0.18 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr1_-_179457805 | 0.18 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr12_-_89920030 | 0.18 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr17_-_33446735 | 0.18 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr17_+_73997796 | 0.17 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr6_-_31846744 | 0.17 |
ENST00000414427.1
ENST00000229729.6 ENST00000375562.4 |
SLC44A4
|
solute carrier family 44, member 4 |
| chr11_-_76155618 | 0.17 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_-_108743471 | 0.17 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr11_-_118135160 | 0.17 |
ENST00000438295.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr12_-_89919965 | 0.17 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr11_-_111794446 | 0.17 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr12_+_49717019 | 0.17 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr11_+_34654011 | 0.17 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr1_+_207277590 | 0.17 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr14_-_94759595 | 0.17 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr11_+_100862811 | 0.16 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr12_-_49523896 | 0.16 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr4_+_119199904 | 0.16 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr3_+_42850959 | 0.16 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr19_+_42055879 | 0.15 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chrX_-_154060946 | 0.15 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr5_-_133304473 | 0.15 |
ENST00000231512.3
|
C5orf15
|
chromosome 5 open reading frame 15 |
| chr1_+_198189921 | 0.15 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr17_-_45056606 | 0.15 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr6_+_147830362 | 0.15 |
ENST00000566741.1
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr9_-_117568365 | 0.15 |
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr3_-_129147432 | 0.15 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr12_-_53901266 | 0.14 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr7_-_92219698 | 0.14 |
ENST00000438306.1
ENST00000445716.1 |
FAM133B
|
family with sequence similarity 133, member B |
| chr9_-_27005686 | 0.14 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr2_-_55496344 | 0.14 |
ENST00000403721.1
ENST00000263629.4 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr8_+_76452097 | 0.14 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_156297919 | 0.14 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr4_+_159131596 | 0.14 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr2_-_18770812 | 0.13 |
ENST00000359846.2
ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
| chr17_-_4938712 | 0.13 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chrX_+_101906294 | 0.13 |
ENST00000361600.5
ENST00000415986.1 ENST00000444152.1 ENST00000537097.1 |
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr10_+_48359344 | 0.13 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr19_-_3600549 | 0.13 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr16_+_16481306 | 0.13 |
ENST00000422673.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr16_+_84402098 | 0.13 |
ENST00000262429.4
ENST00000416219.2 |
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2 |
| chr7_+_141490017 | 0.13 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr15_+_75041184 | 0.13 |
ENST00000343932.4
|
CYP1A2
|
cytochrome P450, family 1, subfamily A, polypeptide 2 |
| chr7_+_95171457 | 0.13 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr6_-_32160622 | 0.12 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr3_+_187957646 | 0.12 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_16685756 | 0.12 |
ENST00000415365.1
ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr1_-_171711387 | 0.12 |
ENST00000236192.7
|
VAMP4
|
vesicle-associated membrane protein 4 |
| chr10_+_104613980 | 0.12 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr2_+_58134756 | 0.12 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr8_+_95835438 | 0.11 |
ENST00000521860.1
ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8
|
integrator complex subunit 8 |
| chr9_+_6645887 | 0.11 |
ENST00000413145.1
|
RP11-390F4.6
|
RP11-390F4.6 |
| chr16_+_22524379 | 0.11 |
ENST00000536620.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr11_+_58874658 | 0.11 |
ENST00000411426.1
|
FAM111B
|
family with sequence similarity 111, member B |
| chr16_-_15736345 | 0.11 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr5_+_70220768 | 0.11 |
ENST00000351205.4
ENST00000503079.2 ENST00000380707.4 ENST00000514951.1 ENST00000506163.1 |
SMN1
|
survival of motor neuron 1, telomeric |
| chr1_+_17531614 | 0.11 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr5_-_141392538 | 0.11 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr8_-_83589388 | 0.11 |
ENST00000522776.1
|
RP11-653B10.1
|
RP11-653B10.1 |
| chr3_-_196669298 | 0.11 |
ENST00000411704.1
ENST00000452404.2 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr2_-_207630033 | 0.11 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr17_+_43213004 | 0.11 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr11_+_125365110 | 0.11 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr8_+_35649365 | 0.11 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr11_-_82782952 | 0.11 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_198299726 | 0.11 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr4_-_156298087 | 0.11 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr2_+_161993412 | 0.10 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr2_-_187367356 | 0.10 |
ENST00000595956.1
|
AC018867.2
|
AC018867.2 |
| chr2_+_207630081 | 0.10 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr19_+_57999101 | 0.10 |
ENST00000347466.6
ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419
|
zinc finger protein 419 |
| chr5_-_143550241 | 0.10 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr17_+_75315654 | 0.10 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chrX_+_72064876 | 0.10 |
ENST00000373532.3
ENST00000333826.5 |
DMRTC1B
|
DMRT-like family C1B |
| chr8_-_29120604 | 0.10 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr17_-_8113542 | 0.10 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr7_+_151771377 | 0.10 |
ENST00000434507.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr12_-_111395610 | 0.10 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chr19_+_56116771 | 0.10 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr19_-_43702231 | 0.10 |
ENST00000597374.1
ENST00000599371.1 |
PSG4
|
pregnancy specific beta-1-glycoprotein 4 |
| chr6_+_28048753 | 0.10 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr12_+_70574088 | 0.10 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr1_-_220220000 | 0.10 |
ENST00000366923.3
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr3_+_141144954 | 0.10 |
ENST00000441582.2
ENST00000321464.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_186265399 | 0.09 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr8_-_66546373 | 0.09 |
ENST00000518908.1
ENST00000458464.2 ENST00000519352.1 |
ARMC1
|
armadillo repeat containing 1 |
| chr7_-_123198284 | 0.09 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr7_+_139026057 | 0.09 |
ENST00000541515.3
|
LUC7L2
|
LUC7-like 2 (S. cerevisiae) |
| chr4_-_156297949 | 0.09 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr10_-_43762329 | 0.09 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr1_-_156647189 | 0.09 |
ENST00000368223.3
|
NES
|
nestin |
| chr20_+_34824355 | 0.09 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr16_+_29832634 | 0.09 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr13_-_47471155 | 0.09 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr6_-_35656685 | 0.09 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr4_-_83719884 | 0.09 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr1_-_12908578 | 0.09 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr10_-_6622258 | 0.09 |
ENST00000263125.5
|
PRKCQ
|
protein kinase C, theta |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 2.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:1903717 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.1 | 0.2 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 1.4 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 1.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.9 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.3 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.4 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.2 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.3 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.3 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) azole transporter activity(GO:0045118) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |