Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
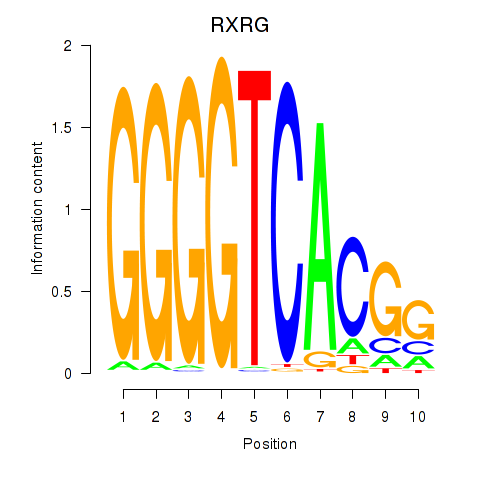
Results for RXRG
Z-value: 2.44
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | retinoid X receptor gamma |
Activity profile of RXRG motif
Sorted Z-values of RXRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_66287257 | 1.97 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr19_-_1174226 | 1.96 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr11_-_615570 | 1.87 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr15_+_41221536 | 1.61 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr19_-_49362621 | 1.55 |
ENST00000594195.1
ENST00000595867.1 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr11_-_615942 | 1.43 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr1_+_23037323 | 1.33 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr12_+_54378849 | 1.22 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr17_+_7210898 | 1.17 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr17_-_66287350 | 1.11 |
ENST00000580666.1
ENST00000583477.1 |
SLC16A6
|
solute carrier family 16, member 6 |
| chr11_+_1093318 | 1.10 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chrX_+_48687283 | 1.06 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr6_-_35888858 | 0.99 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr17_-_27278445 | 0.93 |
ENST00000268756.3
ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr5_-_172755056 | 0.92 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr10_-_79397547 | 0.92 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr13_+_114462193 | 0.92 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr18_-_19748331 | 0.91 |
ENST00000584201.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr17_+_7748233 | 0.91 |
ENST00000570632.1
|
KDM6B
|
lysine (K)-specific demethylase 6B |
| chr5_-_77656175 | 0.89 |
ENST00000513755.1
ENST00000421004.3 |
CTD-2037K23.2
|
CTD-2037K23.2 |
| chr9_+_137533615 | 0.88 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr19_-_1155118 | 0.87 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr22_+_30821732 | 0.86 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr1_-_6420737 | 0.85 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr1_-_212873267 | 0.85 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr18_-_19748379 | 0.84 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr17_+_7184986 | 0.84 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr3_-_120169828 | 0.83 |
ENST00000424703.2
ENST00000469005.1 |
FSTL1
|
follistatin-like 1 |
| chr22_-_51016433 | 0.81 |
ENST00000405237.3
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr20_+_43343886 | 0.81 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_+_47075023 | 0.81 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr20_-_44539538 | 0.80 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr19_-_10047219 | 0.79 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr19_-_4124079 | 0.78 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr20_-_4055812 | 0.78 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr21_-_45196326 | 0.76 |
ENST00000291568.5
|
CSTB
|
cystatin B (stefin B) |
| chr9_-_132515302 | 0.75 |
ENST00000340607.4
|
PTGES
|
prostaglandin E synthase |
| chr19_+_18682531 | 0.73 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr10_-_79397740 | 0.73 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_122399444 | 0.73 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chrX_-_9001351 | 0.73 |
ENST00000362066.3
|
FAM9B
|
family with sequence similarity 9, member B |
| chr11_-_124632179 | 0.71 |
ENST00000278927.5
ENST00000442070.2 ENST00000444566.1 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr17_+_36905613 | 0.70 |
ENST00000539023.1
|
CTB-58E17.5
|
Uncharacterized protein; cDNA FLJ52623 |
| chr19_-_821931 | 0.70 |
ENST00000359894.2
ENST00000520876.3 ENST00000519502.1 |
LPPR3
|
hsa-mir-3187 |
| chr6_+_126112001 | 0.70 |
ENST00000392477.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr4_-_48082192 | 0.68 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr7_+_143078379 | 0.68 |
ENST00000449630.1
ENST00000457235.1 |
ZYX
|
zyxin |
| chr1_-_153521714 | 0.68 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr11_-_104905840 | 0.68 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr12_-_120806960 | 0.67 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr3_-_48672859 | 0.65 |
ENST00000395550.2
ENST00000455886.2 ENST00000431739.1 ENST00000426599.1 ENST00000383733.3 ENST00000420764.2 ENST00000337000.8 |
SLC26A6
|
solute carrier family 26 (anion exchanger), member 6 |
| chr19_-_14228541 | 0.65 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr6_+_31916733 | 0.65 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr11_+_450255 | 0.65 |
ENST00000308020.5
|
PTDSS2
|
phosphatidylserine synthase 2 |
| chr19_+_50354462 | 0.64 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr12_+_52626898 | 0.64 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr17_+_7211280 | 0.63 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr17_-_73844722 | 0.63 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr17_-_47308128 | 0.63 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr22_+_19705928 | 0.63 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chrX_-_49042778 | 0.63 |
ENST00000538114.1
ENST00000376310.3 ENST00000376317.3 ENST00000417014.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr2_+_233243233 | 0.63 |
ENST00000392027.2
|
ALPP
|
alkaline phosphatase, placental |
| chr19_-_56632592 | 0.62 |
ENST00000587279.1
ENST00000270459.3 |
ZNF787
|
zinc finger protein 787 |
| chr16_-_29466285 | 0.61 |
ENST00000330978.3
|
BOLA2
|
bolA family member 2 |
| chr3_+_122399697 | 0.61 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr18_-_21242774 | 0.60 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr6_-_150390202 | 0.60 |
ENST00000438272.2
ENST00000367339.2 |
ULBP3
|
UL16 binding protein 3 |
| chr17_-_56065540 | 0.59 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr15_-_41408409 | 0.59 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr19_+_4402659 | 0.59 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr6_+_32006159 | 0.59 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr19_+_50354393 | 0.58 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr19_+_18682661 | 0.58 |
ENST00000596273.1
ENST00000442744.2 ENST00000595683.1 ENST00000599256.1 ENST00000595158.1 ENST00000598780.1 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr1_+_26606608 | 0.57 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr9_+_131902346 | 0.57 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_-_31840130 | 0.57 |
ENST00000558569.1
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr5_-_131562935 | 0.56 |
ENST00000379104.2
ENST00000379100.2 ENST00000428369.1 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr7_+_100136811 | 0.56 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr15_-_43212996 | 0.56 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr1_-_155880672 | 0.56 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr17_-_43045439 | 0.56 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr15_-_81616446 | 0.55 |
ENST00000302824.6
|
STARD5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr5_+_170846640 | 0.55 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr19_-_39322497 | 0.54 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr22_+_31489344 | 0.54 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr15_+_67430339 | 0.54 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr12_-_54813229 | 0.54 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr19_+_677885 | 0.53 |
ENST00000591552.2
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr12_-_107168696 | 0.53 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr11_-_133826852 | 0.52 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr11_+_65122216 | 0.52 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr1_+_230203010 | 0.52 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chrX_-_132549506 | 0.52 |
ENST00000370828.3
|
GPC4
|
glypican 4 |
| chr10_+_81065975 | 0.52 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr8_+_61822605 | 0.51 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr1_+_113217345 | 0.51 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr15_-_41408339 | 0.51 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr20_-_56285595 | 0.51 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr19_-_18548962 | 0.51 |
ENST00000317018.6
ENST00000581800.1 ENST00000583534.1 ENST00000457269.4 ENST00000338128.8 |
ISYNA1
|
inositol-3-phosphate synthase 1 |
| chr5_-_149682447 | 0.51 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr22_+_19701985 | 0.50 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr9_-_16705069 | 0.49 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr11_+_18417948 | 0.49 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chr12_-_54779511 | 0.49 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_113217043 | 0.49 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr20_+_49411431 | 0.49 |
ENST00000358791.5
ENST00000262591.5 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr20_-_56284816 | 0.49 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr19_+_676385 | 0.49 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr19_+_50353944 | 0.49 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr11_-_59436453 | 0.48 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr9_-_16870704 | 0.48 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr6_-_2971494 | 0.48 |
ENST00000380539.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr7_-_47621229 | 0.48 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr17_-_1531635 | 0.48 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr14_+_103243813 | 0.48 |
ENST00000560371.1
ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3
|
TNF receptor-associated factor 3 |
| chr1_-_202113805 | 0.48 |
ENST00000272217.2
|
ARL8A
|
ADP-ribosylation factor-like 8A |
| chr3_-_197463590 | 0.47 |
ENST00000296343.5
ENST00000389665.5 ENST00000449205.1 |
KIAA0226
|
KIAA0226 |
| chr19_-_39322299 | 0.47 |
ENST00000601094.1
ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr1_+_113217073 | 0.47 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr14_+_24584508 | 0.47 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr15_+_67458357 | 0.47 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr3_-_110612323 | 0.47 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr22_+_51176624 | 0.47 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr5_-_131563501 | 0.47 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr1_-_154946792 | 0.47 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr20_-_49639612 | 0.46 |
ENST00000396017.3
ENST00000433903.1 |
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr1_+_44440575 | 0.46 |
ENST00000532642.1
ENST00000236067.4 ENST00000471859.2 |
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chr19_-_50432782 | 0.46 |
ENST00000413454.1
ENST00000596437.1 ENST00000341114.3 ENST00000595948.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chr2_-_230786679 | 0.46 |
ENST00000543084.1
ENST00000343290.5 ENST00000389044.4 ENST00000283943.5 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr16_+_55357672 | 0.46 |
ENST00000290552.7
|
IRX6
|
iroquois homeobox 6 |
| chr19_+_19516561 | 0.46 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr19_-_50432654 | 0.45 |
ENST00000596680.1
ENST00000594673.1 ENST00000597029.1 |
NUP62
|
nucleoporin 62kDa |
| chr12_-_120805872 | 0.45 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr3_+_14989076 | 0.45 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr19_+_38307999 | 0.45 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr17_-_1532106 | 0.45 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr9_+_135037334 | 0.45 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr22_+_19702069 | 0.44 |
ENST00000412544.1
|
SEPT5
|
septin 5 |
| chr19_+_41094612 | 0.44 |
ENST00000595726.1
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr19_+_54606145 | 0.44 |
ENST00000485876.1
ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr19_+_50354430 | 0.44 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr22_-_41682172 | 0.44 |
ENST00000356244.3
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr6_-_33267101 | 0.44 |
ENST00000497454.1
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr17_-_80291818 | 0.44 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr10_-_79397391 | 0.44 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_167369612 | 0.43 |
ENST00000507747.1
|
RP11-514O12.4
|
RP11-514O12.4 |
| chr1_-_153521597 | 0.43 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr18_-_21242833 | 0.43 |
ENST00000586087.1
ENST00000592179.1 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr7_+_143078652 | 0.43 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr6_+_53659746 | 0.43 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr17_-_27278304 | 0.42 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr19_+_532049 | 0.42 |
ENST00000606136.1
|
CDC34
|
cell division cycle 34 |
| chr19_+_48898132 | 0.42 |
ENST00000263269.3
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr2_+_97203082 | 0.42 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr4_-_140223614 | 0.42 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_-_161015752 | 0.41 |
ENST00000435396.1
ENST00000368021.3 |
USF1
|
upstream transcription factor 1 |
| chr1_+_113217309 | 0.41 |
ENST00000544796.1
ENST00000369644.1 |
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr19_+_18303992 | 0.41 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr7_+_143079000 | 0.41 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr11_+_65407331 | 0.41 |
ENST00000527525.1
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr9_-_110251836 | 0.41 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr19_-_46000251 | 0.41 |
ENST00000590526.1
ENST00000344680.4 ENST00000245923.4 |
RTN2
|
reticulon 2 |
| chrX_+_135229600 | 0.40 |
ENST00000370690.3
|
FHL1
|
four and a half LIM domains 1 |
| chr1_+_245133606 | 0.40 |
ENST00000447569.2
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr11_+_63753883 | 0.40 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr19_+_6361754 | 0.40 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr19_-_14586125 | 0.40 |
ENST00000292513.3
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa |
| chr17_-_39216344 | 0.40 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr20_+_49411543 | 0.40 |
ENST00000609336.1
ENST00000445038.1 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chrX_+_149531524 | 0.40 |
ENST00000370401.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr12_-_117175819 | 0.40 |
ENST00000261318.3
ENST00000536380.1 |
C12orf49
|
chromosome 12 open reading frame 49 |
| chr4_-_140223670 | 0.40 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_-_34664612 | 0.39 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr2_-_230786619 | 0.39 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr19_-_1021113 | 0.39 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr19_+_12780512 | 0.39 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr17_-_76921459 | 0.39 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr11_+_8040739 | 0.39 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr14_-_35873856 | 0.38 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr16_+_67880574 | 0.38 |
ENST00000219169.4
|
NUTF2
|
nuclear transport factor 2 |
| chr8_-_93115445 | 0.38 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chrX_+_153672468 | 0.38 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chrX_+_135229559 | 0.38 |
ENST00000394155.2
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_154946825 | 0.38 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr6_-_33266687 | 0.38 |
ENST00000444031.2
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr16_+_2933187 | 0.38 |
ENST00000396958.3
|
FLYWCH2
|
FLYWCH family member 2 |
| chr20_+_49411523 | 0.38 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr17_-_76778339 | 0.37 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr19_-_51504852 | 0.37 |
ENST00000391806.2
ENST00000347619.4 ENST00000291726.7 ENST00000320838.5 |
KLK8
|
kallikrein-related peptidase 8 |
| chr19_-_50432711 | 0.37 |
ENST00000597723.1
ENST00000599788.1 ENST00000596217.1 ENST00000593652.1 ENST00000599567.1 ENST00000600935.1 ENST00000596011.1 ENST00000596022.1 ENST00000597295.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chr11_+_67806467 | 0.37 |
ENST00000265686.3
ENST00000524598.1 ENST00000529657.1 |
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3 |
| chr15_-_44487408 | 0.37 |
ENST00000402883.1
ENST00000417257.1 |
FRMD5
|
FERM domain containing 5 |
| chr16_+_69458428 | 0.37 |
ENST00000512062.1
ENST00000307892.8 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr13_-_41240717 | 0.37 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chrX_-_153707545 | 0.37 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr2_+_74757050 | 0.37 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr11_-_65430554 | 0.37 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr10_+_47658234 | 0.37 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr19_+_7587555 | 0.37 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr19_+_11998584 | 0.36 |
ENST00000429654.2
ENST00000445911.1 ENST00000340180.5 |
ZNF69
|
zinc finger protein 69 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.7 | 2.8 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.4 | 1.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.3 | 1.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.3 | 0.9 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.7 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.2 | 2.7 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 1.3 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 1.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.9 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.2 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.2 | 0.8 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 0.6 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.2 | 0.6 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 0.8 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.2 | 0.2 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.2 | 1.5 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 1.0 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 1.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.5 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 1.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.6 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 0.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.8 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.5 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 1.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.8 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 0.4 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.8 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.8 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.4 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.5 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.3 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.1 | 0.7 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.6 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.9 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 0.3 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.5 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.6 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 1.4 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.1 | 0.3 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 1.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.3 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.4 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.4 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.4 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.2 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 1.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.7 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.9 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.1 | 0.2 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 0.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.3 | GO:0097647 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.0 | 0.8 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.3 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.2 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 1.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.7 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.6 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.3 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.6 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 1.2 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.2 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 2.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.3 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.9 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 1.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.5 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 1.3 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.0 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.5 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.4 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.6 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.2 | 1.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.2 | 0.4 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.2 | 2.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.6 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.5 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.4 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 0.8 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.7 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.6 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.3 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.1 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.8 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 1.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 1.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.8 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 1.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 2.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.8 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 0.6 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.2 | 0.8 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.0 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.8 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0036361 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 2.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.3 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.5 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 2.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.7 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.7 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 1.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 1.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0050659 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 3.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.0 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0032813 | tumor necrosis factor receptor binding(GO:0005164) tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 2.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.2 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 3.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 0.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 2.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |