Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for RORC
Z-value: 0.44
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORC | hg19_v2_chr1_-_151798546_151798590 | -0.50 | 5.0e-01 | Click! |
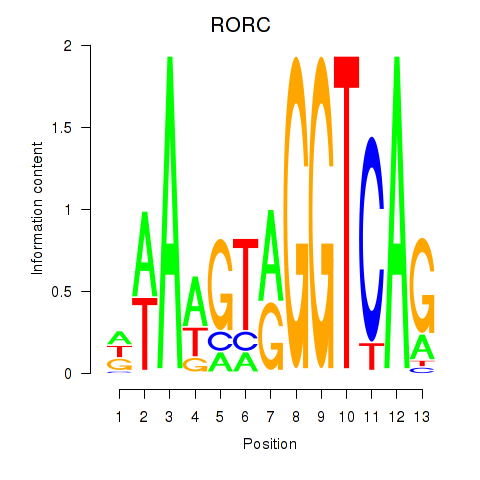
Activity profile of RORC motif
Sorted Z-values of RORC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_39759154 | 0.44 |
ENST00000331982.5
|
IFNL2
|
interferon, lambda 2 |
| chr16_+_1832902 | 0.30 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr12_+_7167980 | 0.28 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr3_-_117716418 | 0.24 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr1_-_1763721 | 0.24 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr4_-_170948361 | 0.22 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_-_8567478 | 0.21 |
ENST00000255612.3
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr12_+_13061894 | 0.20 |
ENST00000540125.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr20_-_33732952 | 0.20 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr11_-_47616210 | 0.19 |
ENST00000302514.3
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4 |
| chr19_-_39735646 | 0.19 |
ENST00000413851.2
|
IFNL3
|
interferon, lambda 3 |
| chr19_+_12780512 | 0.18 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr12_+_93963590 | 0.17 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr17_-_42992856 | 0.15 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr12_+_93964158 | 0.15 |
ENST00000549206.1
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr8_-_116673894 | 0.15 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_+_23025534 | 0.14 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr19_-_46580352 | 0.13 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr22_-_32022280 | 0.13 |
ENST00000442379.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr19_-_7697857 | 0.12 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr1_+_67632083 | 0.12 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr8_+_66955648 | 0.12 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr19_+_38924316 | 0.10 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chrX_-_110655391 | 0.10 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr4_+_110749143 | 0.10 |
ENST00000317735.4
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr12_-_54652060 | 0.10 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr16_+_16326352 | 0.09 |
ENST00000399336.4
ENST00000263012.6 ENST00000538468.1 |
NOMO3
|
NODAL modulator 3 |
| chr2_+_119699864 | 0.09 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr3_-_164914640 | 0.09 |
ENST00000241274.3
|
SLITRK3
|
SLIT and NTRK-like family, member 3 |
| chr9_-_110251836 | 0.09 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr2_+_20650796 | 0.08 |
ENST00000448241.1
|
AC023137.2
|
AC023137.2 |
| chr16_-_1821496 | 0.08 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr14_+_21785693 | 0.08 |
ENST00000382933.4
ENST00000557351.1 |
RPGRIP1
|
retinitis pigmentosa GTPase regulator interacting protein 1 |
| chr6_+_138266498 | 0.08 |
ENST00000434437.1
ENST00000417800.1 |
RP11-240M16.1
|
RP11-240M16.1 |
| chr2_+_119699742 | 0.08 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr4_-_120243545 | 0.08 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr10_-_21435488 | 0.08 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr5_+_177557997 | 0.08 |
ENST00000313386.4
ENST00000515098.1 ENST00000542098.1 ENST00000502814.1 ENST00000507457.1 ENST00000508647.1 |
RMND5B
|
required for meiotic nuclear division 5 homolog B (S. cerevisiae) |
| chr1_+_205197304 | 0.08 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr15_+_31508174 | 0.07 |
ENST00000559292.2
ENST00000557928.1 |
RP11-16E12.1
|
RP11-16E12.1 |
| chr12_+_56414795 | 0.07 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_-_82152444 | 0.07 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr13_+_109248500 | 0.07 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr17_-_48785216 | 0.07 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr6_-_46922659 | 0.06 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chrX_-_43832711 | 0.06 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chrX_-_49089771 | 0.06 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr10_-_105845674 | 0.06 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr10_+_102295616 | 0.06 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr19_+_1269324 | 0.06 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr2_-_30144432 | 0.06 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr1_+_236849754 | 0.05 |
ENST00000542672.1
ENST00000366578.4 |
ACTN2
|
actinin, alpha 2 |
| chr1_-_153522562 | 0.05 |
ENST00000368714.1
|
S100A4
|
S100 calcium binding protein A4 |
| chr10_-_105845536 | 0.05 |
ENST00000393211.3
|
COL17A1
|
collagen, type XVII, alpha 1 |
| chr11_+_65292884 | 0.05 |
ENST00000527009.1
|
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr4_+_140586922 | 0.04 |
ENST00000265498.1
ENST00000506797.1 |
MGST2
|
microsomal glutathione S-transferase 2 |
| chr19_-_10450287 | 0.04 |
ENST00000589261.1
ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3
|
intercellular adhesion molecule 3 |
| chr12_+_4671352 | 0.04 |
ENST00000542744.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_-_217311090 | 0.04 |
ENST00000493603.1
ENST00000366940.2 |
ESRRG
|
estrogen-related receptor gamma |
| chr16_-_67514982 | 0.04 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr3_+_45986511 | 0.04 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr14_-_23755297 | 0.03 |
ENST00000357460.5
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr6_+_44094627 | 0.03 |
ENST00000259746.9
|
TMEM63B
|
transmembrane protein 63B |
| chr3_-_65583561 | 0.03 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr7_+_97736197 | 0.03 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr21_+_17553910 | 0.03 |
ENST00000428669.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_-_44007014 | 0.03 |
ENST00000372726.3
ENST00000537995.1 |
TP53TG5
|
TP53 target 5 |
| chr7_+_127527965 | 0.03 |
ENST00000486037.1
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr4_+_169633310 | 0.03 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_+_20686946 | 0.02 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_-_157069590 | 0.02 |
ENST00000454449.2
|
ETV3L
|
ets variant 3-like |
| chr3_+_160559931 | 0.01 |
ENST00000464260.1
ENST00000295839.9 |
PPM1L
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr16_+_30936971 | 0.01 |
ENST00000565690.1
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_128415844 | 0.01 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr6_+_43738444 | 0.01 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr17_-_10707410 | 0.00 |
ENST00000581851.1
|
LINC00675
|
long intergenic non-protein coding RNA 675 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RORC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |