Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
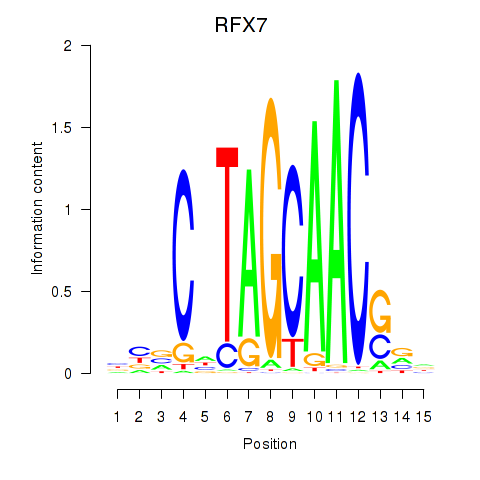
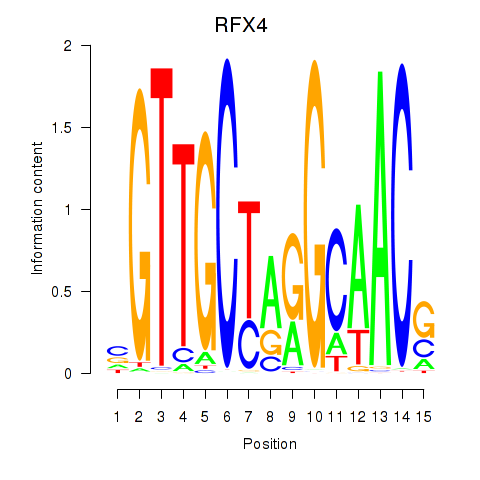
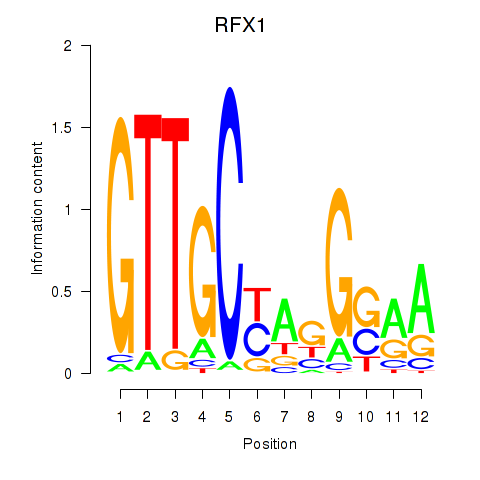
Results for RFX7_RFX4_RFX1
Z-value: 1.54
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | 0.41 | 5.9e-01 | Click! |
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.24 | 7.6e-01 | Click! |
| RFX4 | hg19_v2_chr12_+_106976678_106976708 | -0.19 | 8.1e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51014460 | 1.77 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr19_-_51014588 | 1.62 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr17_+_2264983 | 1.21 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr1_+_161068179 | 1.18 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr19_+_38880695 | 1.07 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr9_-_135545380 | 1.04 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr5_-_7851260 | 0.97 |
ENST00000509627.1
|
C5orf49
|
chromosome 5 open reading frame 49 |
| chr19_-_51014345 | 0.89 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr2_+_54558004 | 0.85 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr17_+_48585794 | 0.76 |
ENST00000576179.1
ENST00000419930.1 |
MYCBPAP
|
MYCBP associated protein |
| chr1_+_43613566 | 0.74 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr16_-_30773372 | 0.74 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr15_-_55790515 | 0.66 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr16_+_24550857 | 0.64 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr4_+_6784358 | 0.63 |
ENST00000508423.1
|
KIAA0232
|
KIAA0232 |
| chr10_+_15001430 | 0.63 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr22_-_50970919 | 0.63 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_-_38607673 | 0.59 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_-_41132088 | 0.56 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr1_+_43613612 | 0.50 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr8_+_94767072 | 0.49 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr8_+_94767109 | 0.49 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr2_-_170550842 | 0.49 |
ENST00000421028.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr5_+_127039075 | 0.47 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr1_-_85155939 | 0.47 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr16_+_30772913 | 0.47 |
ENST00000563909.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr17_-_7108436 | 0.43 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr2_+_170550944 | 0.43 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr1_+_45140360 | 0.42 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr4_+_26585538 | 0.40 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr17_-_55911970 | 0.40 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr8_-_93978309 | 0.40 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr19_-_58609570 | 0.40 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr9_-_117267717 | 0.40 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr7_-_105221898 | 0.39 |
ENST00000486180.1
ENST00000485614.1 ENST00000480514.1 |
EFCAB10
|
EF-hand calcium binding domain 10 |
| chr14_+_89290965 | 0.39 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr16_+_67700673 | 0.39 |
ENST00000403458.4
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr1_+_63989004 | 0.38 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr17_-_45056606 | 0.37 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr21_+_18885318 | 0.37 |
ENST00000400166.1
|
CXADR
|
coxsackie virus and adenovirus receptor |
| chr10_+_82116529 | 0.37 |
ENST00000411538.1
ENST00000256039.2 |
DYDC2
|
DPY30 domain containing 2 |
| chr6_+_46620705 | 0.36 |
ENST00000452689.2
|
SLC25A27
|
solute carrier family 25, member 27 |
| chr2_+_196522032 | 0.36 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr16_-_30064244 | 0.35 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr12_+_109490370 | 0.35 |
ENST00000257548.5
ENST00000536723.1 ENST00000536393.1 |
USP30
|
ubiquitin specific peptidase 30 |
| chr19_+_35634146 | 0.34 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr21_-_43916296 | 0.33 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr14_-_22005062 | 0.33 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_+_29815113 | 0.33 |
ENST00000583755.1
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr4_+_153457404 | 0.33 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr6_-_163148780 | 0.32 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr3_-_48659193 | 0.32 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr22_-_50970506 | 0.32 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_+_814349 | 0.32 |
ENST00000381941.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr14_-_88459182 | 0.31 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr10_+_106113515 | 0.30 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr19_-_11545920 | 0.30 |
ENST00000356392.4
ENST00000591179.1 |
CCDC151
|
coiled-coil domain containing 151 |
| chr1_-_40157345 | 0.30 |
ENST00000372844.3
|
HPCAL4
|
hippocalcin like 4 |
| chr22_-_50970566 | 0.29 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_+_46620676 | 0.29 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr9_+_79792410 | 0.29 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr4_+_26585686 | 0.28 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr9_+_79792269 | 0.28 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr8_+_86999516 | 0.28 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr15_+_49913175 | 0.28 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr2_-_18770812 | 0.27 |
ENST00000359846.2
ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
| chr9_-_4666337 | 0.27 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr4_+_141445333 | 0.27 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr19_-_59084922 | 0.27 |
ENST00000215057.2
ENST00000599369.1 |
MZF1
|
myeloid zinc finger 1 |
| chr15_-_64648273 | 0.27 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr2_+_219187871 | 0.27 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr8_-_144623595 | 0.26 |
ENST00000262577.5
|
ZC3H3
|
zinc finger CCCH-type containing 3 |
| chr2_+_219221573 | 0.26 |
ENST00000289388.3
|
C2orf62
|
chromosome 2 open reading frame 62 |
| chr11_+_71791803 | 0.26 |
ENST00000539271.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr12_+_95611536 | 0.26 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr9_-_140142222 | 0.26 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chrX_-_64254587 | 0.26 |
ENST00000337990.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr6_+_89790459 | 0.26 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr19_-_55791431 | 0.26 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chrX_+_133371077 | 0.25 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr13_+_21141208 | 0.24 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr19_+_19639704 | 0.24 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr19_+_507299 | 0.23 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr9_-_130477912 | 0.23 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr15_+_101142722 | 0.23 |
ENST00000332783.7
ENST00000558747.1 ENST00000343276.4 |
ASB7
|
ankyrin repeat and SOCS box containing 7 |
| chr3_-_128712955 | 0.23 |
ENST00000265068.5
|
KIAA1257
|
KIAA1257 |
| chr6_-_159420780 | 0.23 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr9_-_34381511 | 0.23 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr11_-_64885111 | 0.23 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr9_+_26956474 | 0.23 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr1_+_1981890 | 0.22 |
ENST00000378567.3
ENST00000468310.1 |
PRKCZ
|
protein kinase C, zeta |
| chr12_-_76478386 | 0.22 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr5_+_110074685 | 0.22 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr5_-_122758994 | 0.22 |
ENST00000306467.5
ENST00000515110.1 |
CEP120
|
centrosomal protein 120kDa |
| chr11_+_113185778 | 0.22 |
ENST00000524580.2
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr4_-_140544386 | 0.22 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr11_-_47736896 | 0.22 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr19_+_50321528 | 0.22 |
ENST00000312865.6
ENST00000595185.1 ENST00000538643.1 |
MED25
|
mediator complex subunit 25 |
| chr15_-_101142362 | 0.22 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr22_+_45809560 | 0.22 |
ENST00000342894.3
ENST00000538017.1 |
RIBC2
|
RIB43A domain with coiled-coils 2 |
| chr4_-_156297949 | 0.22 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr1_+_74663994 | 0.21 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr12_-_100660833 | 0.21 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr5_+_121297650 | 0.21 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr1_-_23340400 | 0.21 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr2_-_27851745 | 0.21 |
ENST00000394775.3
ENST00000522876.1 |
CCDC121
|
coiled-coil domain containing 121 |
| chr8_-_93978333 | 0.21 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr20_-_43753104 | 0.21 |
ENST00000372785.3
|
WFDC12
|
WAP four-disulfide core domain 12 |
| chr10_-_82116497 | 0.20 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr2_+_95537248 | 0.20 |
ENST00000427593.2
|
TEKT4
|
tektin 4 |
| chr2_+_202937972 | 0.20 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr14_+_93357749 | 0.20 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr11_+_124543694 | 0.20 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr15_+_49913201 | 0.20 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr11_+_537494 | 0.20 |
ENST00000270115.7
|
LRRC56
|
leucine rich repeat containing 56 |
| chr16_-_67450325 | 0.20 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr2_-_62081254 | 0.20 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr9_+_34458851 | 0.19 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr16_+_19729586 | 0.19 |
ENST00000564186.1
ENST00000541926.1 ENST00000433597.2 |
IQCK
|
IQ motif containing K |
| chr7_+_99006232 | 0.19 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr17_-_15244894 | 0.19 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr21_-_36259445 | 0.19 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr15_-_101142401 | 0.19 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr11_-_66360548 | 0.19 |
ENST00000333861.3
|
CCDC87
|
coiled-coil domain containing 87 |
| chr17_+_48585902 | 0.18 |
ENST00000452039.1
|
MYCBPAP
|
MYCBP associated protein |
| chr1_+_6640046 | 0.18 |
ENST00000319084.5
ENST00000435905.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chrX_+_135251783 | 0.18 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr15_-_49912987 | 0.18 |
ENST00000560246.1
ENST00000558594.1 |
FAM227B
|
family with sequence similarity 227, member B |
| chr6_+_89790490 | 0.18 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr7_+_123485102 | 0.18 |
ENST00000488323.1
ENST00000223026.4 |
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr12_-_76478446 | 0.18 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_-_73882249 | 0.18 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr20_+_48807351 | 0.17 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr2_+_196521845 | 0.17 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr19_-_55791563 | 0.17 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr11_-_8615507 | 0.17 |
ENST00000431279.2
ENST00000418597.1 |
STK33
|
serine/threonine kinase 33 |
| chr3_+_97483572 | 0.17 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr19_-_5680891 | 0.17 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr1_-_207226313 | 0.17 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr10_+_5454505 | 0.17 |
ENST00000355029.4
|
NET1
|
neuroepithelial cell transforming 1 |
| chr20_+_18447771 | 0.17 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr16_-_29757272 | 0.17 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr19_+_10217364 | 0.17 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr19_-_40324767 | 0.17 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr20_+_19997948 | 0.16 |
ENST00000310450.4
ENST00000398602.2 |
NAA20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr15_+_75487984 | 0.16 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr14_-_88459503 | 0.16 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr6_-_159065741 | 0.16 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr16_+_19078911 | 0.16 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr5_+_111964133 | 0.16 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chrX_+_47444613 | 0.16 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr4_+_20702030 | 0.16 |
ENST00000510051.1
ENST00000503585.1 ENST00000360916.5 ENST00000295290.8 ENST00000514485.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr11_+_113185571 | 0.16 |
ENST00000455306.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr13_-_52733670 | 0.16 |
ENST00000550841.1
|
NEK3
|
NIMA-related kinase 3 |
| chr6_+_134274354 | 0.16 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr5_-_68665084 | 0.16 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr16_+_57406437 | 0.16 |
ENST00000564948.1
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr9_-_4666421 | 0.16 |
ENST00000381895.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr12_+_111051832 | 0.16 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr6_-_163148700 | 0.16 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr22_-_32555275 | 0.16 |
ENST00000382097.3
|
C22orf42
|
chromosome 22 open reading frame 42 |
| chr14_-_102771516 | 0.16 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr16_+_2059872 | 0.16 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr3_+_14474178 | 0.15 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr1_+_150480551 | 0.15 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr6_+_43484760 | 0.15 |
ENST00000372389.3
ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa |
| chr3_-_45883558 | 0.15 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr6_+_17281802 | 0.15 |
ENST00000509686.1
|
RBM24
|
RNA binding motif protein 24 |
| chr15_-_72612470 | 0.15 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr10_-_21435488 | 0.15 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr13_-_44735393 | 0.15 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr14_-_102771462 | 0.15 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr7_-_27224795 | 0.15 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr5_-_76916396 | 0.15 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr17_-_6543973 | 0.15 |
ENST00000571642.1
ENST00000572370.1 |
KIAA0753
|
KIAA0753 |
| chr8_+_96281045 | 0.15 |
ENST00000521905.1
|
KB-1047C11.2
|
KB-1047C11.2 |
| chr16_+_15031300 | 0.15 |
ENST00000328085.6
|
NPIPA1
|
nuclear pore complex interacting protein family, member A1 |
| chr14_-_61447752 | 0.15 |
ENST00000555420.1
ENST00000553903.1 |
TRMT5
|
tRNA methyltransferase 5 |
| chr7_-_100808843 | 0.14 |
ENST00000249330.2
|
VGF
|
VGF nerve growth factor inducible |
| chr7_+_23636992 | 0.14 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr16_+_16429787 | 0.14 |
ENST00000331436.4
ENST00000541593.1 |
AC138969.4
|
Protein PKD1P1 |
| chrX_-_131623874 | 0.14 |
ENST00000436215.1
|
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr1_-_45140074 | 0.14 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr1_-_38061522 | 0.14 |
ENST00000373062.3
|
GNL2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr4_+_123653807 | 0.14 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr14_-_74416829 | 0.14 |
ENST00000534936.1
|
FAM161B
|
family with sequence similarity 161, member B |
| chr11_+_61717535 | 0.14 |
ENST00000534553.1
ENST00000301774.9 |
BEST1
|
bestrophin 1 |
| chr1_+_40627038 | 0.14 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr11_+_85339623 | 0.14 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chrX_+_135252050 | 0.14 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr15_-_49103235 | 0.14 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr19_-_51523412 | 0.14 |
ENST00000391805.1
ENST00000599077.1 |
KLK10
|
kallikrein-related peptidase 10 |
| chr8_-_93978346 | 0.14 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr5_-_143550241 | 0.14 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr6_+_134274322 | 0.14 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr9_-_72435576 | 0.13 |
ENST00000453410.1
ENST00000526458.1 ENST00000439418.1 |
C9orf135-AS1
|
C9orf135 antisense RNA 1 (head to head) |
| chr7_-_100808394 | 0.13 |
ENST00000445482.2
|
VGF
|
VGF nerve growth factor inducible |
| chr6_+_135818907 | 0.13 |
ENST00000579339.1
ENST00000580741.1 |
LINC00271
|
long intergenic non-protein coding RNA 271 |
| chr7_-_123174610 | 0.13 |
ENST00000324698.6
ENST00000434450.1 |
IQUB
|
IQ motif and ubiquitin domain containing |
| chr4_+_141445311 | 0.13 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.6 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.6 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.3 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.4 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.3 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.7 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.2 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.1 | 0.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.2 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 0.2 | GO:0039023 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.4 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.2 | GO:2001176 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.2 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 1.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.4 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.4 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.3 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:1902361 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.3 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.3 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.4 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 4.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |