Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
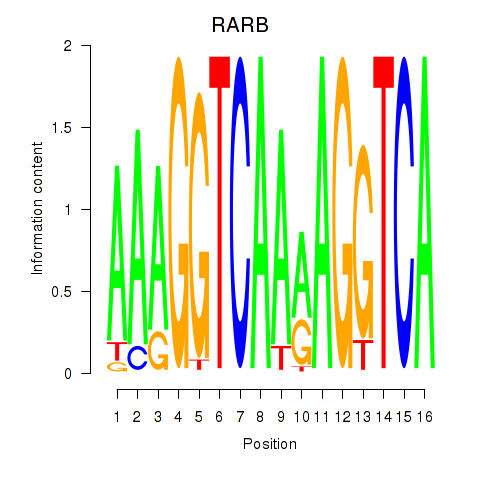
Results for RARB
Z-value: 1.26
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.14 | retinoic acid receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARB | hg19_v2_chr3_+_25469724_25469773 | 0.98 | 2.3e-02 | Click! |
Activity profile of RARB motif
Sorted Z-values of RARB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_100140331 | 1.93 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr9_+_67977438 | 1.88 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr7_-_98805129 | 1.35 |
ENST00000327442.6
|
KPNA7
|
karyopherin alpha 7 (importin alpha 8) |
| chr6_+_49431073 | 1.14 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr3_+_119316721 | 1.10 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr22_-_37976082 | 1.02 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr6_-_49430886 | 1.02 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr10_-_101825151 | 0.96 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr8_-_95274536 | 0.91 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr3_+_119316689 | 0.91 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr15_-_31521567 | 0.89 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr17_-_2318731 | 0.88 |
ENST00000609667.1
|
AC006435.1
|
Uncharacterized protein |
| chr19_+_29704142 | 0.86 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr22_+_39493207 | 0.82 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr19_-_58204128 | 0.80 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr15_+_29211570 | 0.79 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr2_+_217082311 | 0.77 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr17_-_41277317 | 0.75 |
ENST00000497488.1
ENST00000489037.1 ENST00000470026.1 ENST00000586385.1 ENST00000591534.1 ENST00000591849.1 |
BRCA1
|
breast cancer 1, early onset |
| chr8_+_74903580 | 0.67 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr19_-_9731872 | 0.65 |
ENST00000424629.1
ENST00000326044.5 ENST00000354661.4 ENST00000435550.1 ENST00000444611.1 ENST00000421525.1 |
ZNF561
|
zinc finger protein 561 |
| chr17_-_41277370 | 0.65 |
ENST00000476777.1
ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1
|
breast cancer 1, early onset |
| chr18_+_657578 | 0.64 |
ENST00000323274.10
|
TYMS
|
thymidylate synthetase |
| chr19_+_21106081 | 0.64 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr9_+_104296163 | 0.63 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chrX_+_23682379 | 0.58 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr12_-_112443830 | 0.57 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr1_+_35544968 | 0.56 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr1_+_53392901 | 0.56 |
ENST00000371514.3
ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2
|
sterol carrier protein 2 |
| chr6_-_52859046 | 0.54 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr4_+_56719782 | 0.53 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr1_-_109618566 | 0.53 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr13_+_73302047 | 0.52 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr16_-_46649905 | 0.51 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr11_+_95523621 | 0.51 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr4_+_2451700 | 0.51 |
ENST00000506607.1
|
RP11-503N18.3
|
Uncharacterized protein |
| chrY_-_23548246 | 0.50 |
ENST00000382764.1
|
CYorf17
|
chromosome Y open reading frame 17 |
| chr1_+_28836561 | 0.49 |
ENST00000419074.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr5_+_27472371 | 0.48 |
ENST00000512067.1
ENST00000510165.1 ENST00000505775.1 ENST00000514844.1 ENST00000503107.1 |
LINC01021
|
long intergenic non-protein coding RNA 1021 |
| chr12_+_20522179 | 0.46 |
ENST00000359062.3
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited |
| chr10_-_27389320 | 0.46 |
ENST00000436985.2
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr3_+_49297518 | 0.45 |
ENST00000440528.3
|
RP11-3B7.1
|
Uncharacterized protein |
| chr12_+_57810198 | 0.42 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr12_+_57854274 | 0.42 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr19_+_38880695 | 0.42 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chrX_-_119709637 | 0.42 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr11_-_798239 | 0.41 |
ENST00000531437.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr6_+_76311568 | 0.41 |
ENST00000370014.3
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr1_-_150693318 | 0.40 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr15_-_54051831 | 0.40 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr1_-_53019059 | 0.39 |
ENST00000484723.2
ENST00000524582.1 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr4_+_128802016 | 0.38 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chrX_+_123095890 | 0.38 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr1_-_86861660 | 0.38 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr3_-_195938256 | 0.37 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr19_+_18485509 | 0.37 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr4_+_124317940 | 0.36 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr6_+_2988199 | 0.36 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chr1_+_52521797 | 0.36 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr10_-_33625154 | 0.35 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr2_-_3521518 | 0.35 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr5_-_141392538 | 0.34 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr7_-_5998714 | 0.34 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr11_-_796197 | 0.34 |
ENST00000530360.1
ENST00000528606.1 ENST00000320230.5 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr12_+_56075330 | 0.34 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr2_-_175499294 | 0.33 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_110052302 | 0.33 |
ENST00000369864.4
ENST00000369862.1 |
AMIGO1
|
adhesion molecule with Ig-like domain 1 |
| chr4_-_171011323 | 0.33 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr5_-_133968529 | 0.32 |
ENST00000402673.2
|
SAR1B
|
SAR1 homolog B (S. cerevisiae) |
| chr1_+_221051699 | 0.31 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr1_-_52521831 | 0.31 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_+_140174429 | 0.29 |
ENST00000520672.2
ENST00000378132.1 ENST00000526136.1 |
PCDHA2
|
protocadherin alpha 2 |
| chr2_+_132287237 | 0.29 |
ENST00000467992.2
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr1_-_150693305 | 0.29 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr6_+_76311736 | 0.29 |
ENST00000447266.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr2_-_25194476 | 0.28 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr12_-_112443767 | 0.28 |
ENST00000550233.1
ENST00000546962.1 ENST00000550800.2 |
TMEM116
|
transmembrane protein 116 |
| chr11_+_31531291 | 0.28 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr10_-_65028938 | 0.27 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr1_+_78245303 | 0.27 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr22_+_39493268 | 0.26 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr11_-_506316 | 0.26 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr10_-_43762329 | 0.26 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr1_+_52521957 | 0.26 |
ENST00000472944.2
ENST00000484036.1 |
BTF3L4
|
basic transcription factor 3-like 4 |
| chr12_+_49658855 | 0.25 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr8_-_141774467 | 0.25 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr20_+_43990576 | 0.25 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr19_+_49468558 | 0.24 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr18_-_21852143 | 0.24 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr6_+_31638156 | 0.24 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr10_-_27530997 | 0.23 |
ENST00000375901.1
ENST00000412279.1 ENST00000375905.4 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr4_-_69536346 | 0.23 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chrX_+_123095860 | 0.23 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr16_-_1275257 | 0.23 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr19_+_13228917 | 0.22 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr16_+_2533020 | 0.22 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr19_-_33360647 | 0.22 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr19_+_5914213 | 0.22 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr6_-_28367510 | 0.22 |
ENST00000361028.1
|
ZSCAN12
|
zinc finger and SCAN domain containing 12 |
| chr10_-_65028817 | 0.21 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr9_-_104145795 | 0.21 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr4_-_7436671 | 0.21 |
ENST00000319098.4
|
PSAPL1
|
prosaposin-like 1 (gene/pseudogene) |
| chr17_+_73997419 | 0.21 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chrX_-_134429952 | 0.20 |
ENST00000370764.1
|
ZNF75D
|
zinc finger protein 75D |
| chr6_+_88106840 | 0.20 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr1_-_150979333 | 0.20 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr1_+_202792385 | 0.19 |
ENST00000549576.1
|
RP11-480I12.4
|
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr2_+_29117509 | 0.19 |
ENST00000407426.3
|
WDR43
|
WD repeat domain 43 |
| chr10_-_8095412 | 0.19 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr6_+_24495067 | 0.19 |
ENST00000357578.3
ENST00000546278.1 ENST00000491546.1 |
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr6_+_31637944 | 0.19 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr9_-_3489406 | 0.18 |
ENST00000457373.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr7_-_141401951 | 0.18 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr11_-_129817471 | 0.18 |
ENST00000423662.2
|
PRDM10
|
PR domain containing 10 |
| chr7_-_56160625 | 0.18 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr17_-_46178527 | 0.18 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr12_-_2966193 | 0.18 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr6_+_147527103 | 0.18 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr6_-_33771757 | 0.18 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr7_-_107204918 | 0.17 |
ENST00000297135.3
|
COG5
|
component of oligomeric golgi complex 5 |
| chr1_+_59775752 | 0.17 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr2_-_107084826 | 0.17 |
ENST00000304514.7
ENST00000409886.3 |
RGPD3
|
RANBP2-like and GRIP domain containing 3 |
| chr11_+_111783450 | 0.17 |
ENST00000537382.1
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr5_-_74162605 | 0.16 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr10_-_81203972 | 0.16 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr1_+_75594119 | 0.16 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr6_-_31763721 | 0.16 |
ENST00000375663.3
|
VARS
|
valyl-tRNA synthetase |
| chr6_-_24667180 | 0.16 |
ENST00000545995.1
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr3_-_121448791 | 0.16 |
ENST00000489400.1
|
GOLGB1
|
golgin B1 |
| chr22_-_21360736 | 0.16 |
ENST00000547793.2
|
AC002472.1
|
Uncharacterized protein |
| chr19_-_15344243 | 0.15 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr14_+_89290965 | 0.15 |
ENST00000345383.5
ENST00000536576.1 ENST00000346301.4 ENST00000338104.6 ENST00000354441.6 ENST00000380656.2 ENST00000556651.1 ENST00000554686.1 |
TTC8
|
tetratricopeptide repeat domain 8 |
| chr4_+_8594477 | 0.15 |
ENST00000315782.6
|
CPZ
|
carboxypeptidase Z |
| chr14_+_104710541 | 0.15 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr11_-_111782484 | 0.15 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr11_-_116708302 | 0.15 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr11_-_95522907 | 0.15 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr3_+_99986036 | 0.14 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr2_-_152830479 | 0.14 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_-_75008244 | 0.14 |
ENST00000510798.1
ENST00000446329.2 |
POC5
|
POC5 centriolar protein |
| chr4_-_84406218 | 0.14 |
ENST00000515303.1
|
FAM175A
|
family with sequence similarity 175, member A |
| chr17_-_46178741 | 0.14 |
ENST00000581003.1
ENST00000225603.4 |
CBX1
|
chromobox homolog 1 |
| chr22_+_24819565 | 0.14 |
ENST00000424232.1
ENST00000486108.1 |
ADORA2A
|
adenosine A2a receptor |
| chr16_-_18923035 | 0.14 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr17_-_40828969 | 0.14 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_+_41363854 | 0.13 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr1_+_174669653 | 0.13 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr17_+_58677539 | 0.13 |
ENST00000305921.3
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr17_-_40829026 | 0.13 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr6_-_24667232 | 0.13 |
ENST00000378198.4
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr9_+_104296243 | 0.13 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr17_-_28618948 | 0.13 |
ENST00000261714.6
|
BLMH
|
bleomycin hydrolase |
| chr10_+_11784360 | 0.13 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr4_+_77177512 | 0.13 |
ENST00000606246.1
|
FAM47E
|
family with sequence similarity 47, member E |
| chr6_+_168434678 | 0.13 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chrX_-_14047996 | 0.12 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr6_+_76311636 | 0.12 |
ENST00000327284.8
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr7_-_74489609 | 0.12 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr7_-_123198284 | 0.12 |
ENST00000355749.2
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr2_+_88991162 | 0.12 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chr22_-_43398442 | 0.12 |
ENST00000422336.1
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr14_-_80697396 | 0.11 |
ENST00000557010.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr8_+_91013676 | 0.11 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr5_-_135290651 | 0.11 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr11_+_63870660 | 0.11 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr17_+_76126842 | 0.11 |
ENST00000590426.1
ENST00000590799.1 ENST00000318430.5 ENST00000589691.1 |
TMC8
|
transmembrane channel-like 8 |
| chr11_+_118398178 | 0.11 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chrX_+_56100757 | 0.11 |
ENST00000433279.1
|
AL353698.1
|
Uncharacterized protein |
| chr11_+_111782934 | 0.10 |
ENST00000304298.3
|
HSPB2
|
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chrX_+_73164167 | 0.10 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr8_+_13424352 | 0.10 |
ENST00000297324.4
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr8_-_27462822 | 0.10 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr11_+_61520075 | 0.10 |
ENST00000278836.5
|
MYRF
|
myelin regulatory factor |
| chr18_-_32924372 | 0.10 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr16_-_21875424 | 0.10 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr1_+_17944832 | 0.10 |
ENST00000167825.4
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr14_+_62164340 | 0.09 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr4_+_8594435 | 0.09 |
ENST00000382480.2
|
CPZ
|
carboxypeptidase Z |
| chr9_-_125590818 | 0.09 |
ENST00000259467.4
|
PDCL
|
phosducin-like |
| chr19_-_11688260 | 0.09 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr1_+_150898733 | 0.09 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr9_-_100459639 | 0.09 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr11_-_62476694 | 0.08 |
ENST00000524862.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr17_+_26800756 | 0.08 |
ENST00000537681.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr20_-_5485166 | 0.08 |
ENST00000589201.1
ENST00000379053.4 |
LINC00654
|
long intergenic non-protein coding RNA 654 |
| chr17_+_26800648 | 0.08 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr8_+_8860314 | 0.08 |
ENST00000250263.7
ENST00000519292.1 |
ERI1
|
exoribonuclease 1 |
| chr3_-_49837254 | 0.08 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr12_-_323248 | 0.08 |
ENST00000535347.1
ENST00000536824.1 |
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr4_+_8594364 | 0.08 |
ENST00000360986.4
|
CPZ
|
carboxypeptidase Z |
| chr6_-_31763408 | 0.08 |
ENST00000444930.2
|
VARS
|
valyl-tRNA synthetase |
| chr3_-_64009658 | 0.08 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr5_+_147774275 | 0.08 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr10_-_61899124 | 0.08 |
ENST00000373815.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr16_+_2106134 | 0.07 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr17_+_78518617 | 0.07 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr11_-_798305 | 0.07 |
ENST00000531514.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr12_-_76377795 | 0.07 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chrX_+_73164149 | 0.07 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chrX_+_102024075 | 0.07 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr5_-_74162739 | 0.07 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.3 | 1.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 0.9 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.2 | 1.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.6 | GO:0019860 | uracil metabolic process(GO:0019860) |
| 0.2 | 1.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.6 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.8 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.3 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.4 | GO:0060032 | notochord regression(GO:0060032) |
| 0.1 | 1.8 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 0.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.8 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.6 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 1.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 0.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.4 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 1.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.2 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.0 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.5 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 1.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 1.4 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.6 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.2 | 0.6 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.2 | 0.5 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.3 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.1 | 0.3 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.8 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.4 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 1.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |