Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PTF1A
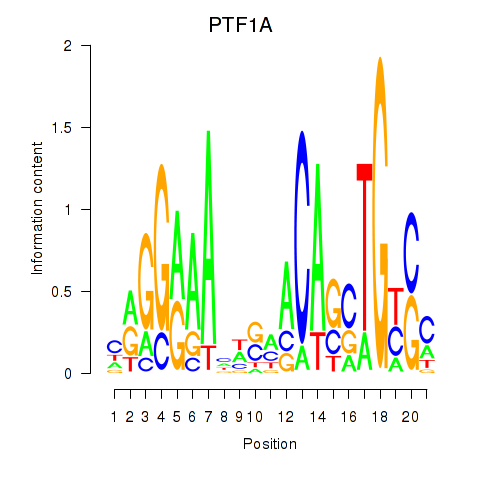
Z-value: 1.43
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113344811 | 1.65 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_113344582 | 1.44 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_113344755 | 1.37 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_23037323 | 1.33 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr4_-_129491686 | 1.29 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr2_-_106777040 | 1.13 |
ENST00000444193.1
ENST00000416298.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr3_-_172241250 | 1.12 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr1_+_150480551 | 1.07 |
ENST00000369049.4
ENST00000369047.4 |
ECM1
|
extracellular matrix protein 1 |
| chr12_+_2912363 | 1.03 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr1_+_150480576 | 0.87 |
ENST00000346569.6
|
ECM1
|
extracellular matrix protein 1 |
| chr10_-_10504285 | 0.77 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr19_+_29493486 | 0.73 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr15_+_41099919 | 0.65 |
ENST00000561617.1
|
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr18_+_33877654 | 0.64 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr19_+_11670245 | 0.61 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr15_+_41099788 | 0.61 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr2_-_1748214 | 0.60 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr6_-_46620522 | 0.58 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr14_+_67656110 | 0.57 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr17_+_67498295 | 0.56 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_-_61735103 | 0.54 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr8_+_32405728 | 0.52 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr5_-_173217931 | 0.50 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr2_-_239140011 | 0.50 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr12_+_75784850 | 0.49 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr12_+_112451222 | 0.49 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr1_-_205649580 | 0.48 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr20_+_42544782 | 0.48 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr8_+_32405785 | 0.45 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr6_-_134499037 | 0.44 |
ENST00000528577.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_+_101546827 | 0.44 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr19_-_44258733 | 0.43 |
ENST00000597586.1
ENST00000596714.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr17_-_61778528 | 0.42 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chr16_+_2802623 | 0.42 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr19_+_54496132 | 0.41 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr1_-_9129598 | 0.41 |
ENST00000535586.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr11_-_45687128 | 0.40 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr17_+_79859985 | 0.39 |
ENST00000333383.7
|
NPB
|
neuropeptide B |
| chr2_-_196933536 | 0.38 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr4_+_81951957 | 0.36 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr3_-_88108212 | 0.36 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_-_27435125 | 0.36 |
ENST00000414408.1
ENST00000310574.3 |
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr19_+_29493456 | 0.36 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chrX_+_15808569 | 0.35 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr19_-_17014408 | 0.34 |
ENST00000594249.1
|
CPAMD8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chrX_+_48242863 | 0.34 |
ENST00000376886.2
ENST00000375517.3 |
SSX4
|
synovial sarcoma, X breakpoint 4 |
| chr14_-_35099315 | 0.34 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr15_+_68924327 | 0.34 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr8_-_135725205 | 0.33 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr1_+_178694300 | 0.32 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_-_123843597 | 0.32 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr15_+_91446961 | 0.32 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr12_+_31079652 | 0.32 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr8_-_145641864 | 0.32 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr21_+_44073916 | 0.32 |
ENST00000349112.3
ENST00000398224.3 |
PDE9A
|
phosphodiesterase 9A |
| chr17_+_68047418 | 0.31 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr22_-_50523760 | 0.30 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_-_12883026 | 0.29 |
ENST00000396953.2
ENST00000457131.1 ENST00000435983.1 ENST00000273223.6 ENST00000396957.1 ENST00000429711.2 |
RPL32
|
ribosomal protein L32 |
| chr21_+_44073860 | 0.29 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chr15_+_84841242 | 0.29 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr11_+_71276609 | 0.29 |
ENST00000398531.1
ENST00000376536.4 |
KRTAP5-10
|
keratin associated protein 5-10 |
| chr11_-_64052111 | 0.29 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr16_+_2802316 | 0.28 |
ENST00000301740.8
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr2_-_228497888 | 0.28 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr17_-_33415837 | 0.28 |
ENST00000414419.2
|
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr17_-_73892992 | 0.27 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr19_+_45254529 | 0.27 |
ENST00000444487.1
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr10_+_43572475 | 0.27 |
ENST00000355710.3
ENST00000498820.1 ENST00000340058.5 |
RET
|
ret proto-oncogene |
| chr2_-_86422523 | 0.27 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr1_+_178694362 | 0.26 |
ENST00000367634.2
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_-_2962331 | 0.26 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr15_+_81225699 | 0.26 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr6_-_62996066 | 0.25 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr1_+_157963391 | 0.25 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr8_-_145642203 | 0.24 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_+_40204538 | 0.24 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr19_+_54495542 | 0.24 |
ENST00000252729.2
ENST00000352529.1 |
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr10_-_28571015 | 0.23 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_9129895 | 0.23 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_-_22089533 | 0.21 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr2_+_173600565 | 0.21 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_35099377 | 0.21 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr19_-_48389651 | 0.20 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr17_+_12692774 | 0.20 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr12_+_112451120 | 0.20 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr2_+_173600514 | 0.20 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr8_-_110660975 | 0.19 |
ENST00000528045.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr4_-_87278857 | 0.18 |
ENST00000509464.1
ENST00000511167.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_+_85922491 | 0.18 |
ENST00000526018.1
|
GNLY
|
granulysin |
| chr11_+_64052266 | 0.18 |
ENST00000539851.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr2_-_71454185 | 0.18 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr2_+_120187465 | 0.18 |
ENST00000409826.1
ENST00000417645.1 |
TMEM37
|
transmembrane protein 37 |
| chr12_+_109826524 | 0.17 |
ENST00000431443.2
|
MYO1H
|
myosin IH |
| chr1_+_151682909 | 0.17 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr20_-_48732472 | 0.17 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr8_-_22089845 | 0.17 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr1_-_45956868 | 0.17 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr3_+_35681081 | 0.16 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr19_+_6464502 | 0.16 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr17_+_67498538 | 0.16 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_89744365 | 0.16 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr2_+_3393453 | 0.15 |
ENST00000441983.1
|
TRAPPC12
|
trafficking protein particle complex 12 |
| chrX_-_134305322 | 0.15 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr16_-_65155833 | 0.15 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr12_+_12966250 | 0.15 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr22_-_17640110 | 0.15 |
ENST00000399852.3
ENST00000336737.4 |
CECR5
|
cat eye syndrome chromosome region, candidate 5 |
| chr11_+_64052454 | 0.14 |
ENST00000539833.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr2_-_27434611 | 0.14 |
ENST00000408041.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr14_-_38028689 | 0.14 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr11_+_8704748 | 0.14 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr10_+_43633914 | 0.14 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr6_-_24877490 | 0.14 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr5_+_125800836 | 0.13 |
ENST00000511134.1
|
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_13956497 | 0.13 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr1_-_45956800 | 0.12 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr19_-_1168936 | 0.12 |
ENST00000587655.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr7_+_6714599 | 0.12 |
ENST00000328239.7
ENST00000542006.1 |
AC073343.1
|
Uncharacterized protein |
| chr4_-_89744314 | 0.12 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr17_+_6918064 | 0.12 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr17_-_33416231 | 0.12 |
ENST00000584655.1
ENST00000447669.2 ENST00000315249.7 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr3_-_193272874 | 0.12 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr3_+_40518599 | 0.12 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr1_-_33647267 | 0.11 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr1_+_171454639 | 0.11 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr6_-_127780510 | 0.10 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr8_+_97773457 | 0.10 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr5_+_132149017 | 0.10 |
ENST00000378693.2
|
SOWAHA
|
sosondowah ankyrin repeat domain family member A |
| chr5_-_39425290 | 0.10 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr14_+_70346125 | 0.10 |
ENST00000361956.3
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr18_-_53177984 | 0.10 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr12_+_53689309 | 0.10 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr11_+_64052294 | 0.10 |
ENST00000536667.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr22_-_50051151 | 0.10 |
ENST00000400023.1
ENST00000444628.1 |
C22orf34
|
chromosome 22 open reading frame 34 |
| chr17_-_42143963 | 0.09 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr4_+_40198527 | 0.09 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr4_-_89744457 | 0.09 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr16_+_5121814 | 0.08 |
ENST00000262374.5
ENST00000586840.1 |
ALG1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr13_-_31736132 | 0.08 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr1_-_40782347 | 0.08 |
ENST00000417105.1
|
COL9A2
|
collagen, type IX, alpha 2 |
| chr17_+_67498396 | 0.08 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr6_-_24489842 | 0.08 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr8_+_11351876 | 0.08 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr4_-_175750364 | 0.07 |
ENST00000340217.5
ENST00000274093.3 |
GLRA3
|
glycine receptor, alpha 3 |
| chr6_-_41715128 | 0.07 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr1_+_100503643 | 0.06 |
ENST00000370152.3
|
HIAT1
|
hippocampus abundant transcript 1 |
| chr3_-_169587621 | 0.06 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr5_+_173315283 | 0.06 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr8_+_11351494 | 0.06 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr2_+_27435179 | 0.05 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr4_-_5990166 | 0.05 |
ENST00000324058.5
|
C4orf50
|
chromosome 4 open reading frame 50 |
| chr5_-_39425068 | 0.05 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr21_+_39668831 | 0.05 |
ENST00000419868.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_-_3461092 | 0.05 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr19_-_46526304 | 0.05 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr2_-_237416181 | 0.05 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr2_-_237416071 | 0.05 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr5_-_39425222 | 0.05 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_36499693 | 0.04 |
ENST00000342292.4
|
GPR179
|
G protein-coupled receptor 179 |
| chr11_+_64052692 | 0.04 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr13_-_111522162 | 0.04 |
ENST00000538077.1
|
LINC00346
|
long intergenic non-protein coding RNA 346 |
| chr4_+_184826418 | 0.04 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr3_+_51663407 | 0.04 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr1_+_178694408 | 0.03 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr5_-_16916624 | 0.03 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr11_-_1629693 | 0.03 |
ENST00000399685.1
|
KRTAP5-3
|
keratin associated protein 5-3 |
| chr13_-_79979952 | 0.03 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr9_-_19033185 | 0.03 |
ENST00000380534.4
|
FAM154A
|
family with sequence similarity 154, member A |
| chr9_-_116061145 | 0.02 |
ENST00000416588.2
|
RNF183
|
ring finger protein 183 |
| chr3_-_122712079 | 0.02 |
ENST00000393583.2
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr10_-_74856608 | 0.02 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr16_-_15950868 | 0.02 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr13_+_113812956 | 0.02 |
ENST00000375547.2
ENST00000342783.4 |
PROZ
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr4_-_82393009 | 0.02 |
ENST00000436139.2
|
RASGEF1B
|
RasGEF domain family, member 1B |
| chr16_-_2004683 | 0.02 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr19_-_11039261 | 0.02 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr7_-_72993033 | 0.01 |
ENST00000305632.5
|
TBL2
|
transducin (beta)-like 2 |
| chr11_+_207477 | 0.01 |
ENST00000526104.1
|
RIC8A
|
RIC8 guanine nucleotide exchange factor A |
| chr7_-_99097863 | 0.01 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr20_+_30697298 | 0.01 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr2_-_95831158 | 0.01 |
ENST00000447814.1
|
ZNF514
|
zinc finger protein 514 |
| chr13_-_79979919 | 0.01 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr1_-_9129735 | 0.01 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr22_+_24999114 | 0.01 |
ENST00000412658.1
ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr7_+_150688166 | 0.01 |
ENST00000461406.1
|
NOS3
|
nitric oxide synthase 3 (endothelial cell) |
| chr9_-_138853156 | 0.00 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr1_-_45956822 | 0.00 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr9_+_4985228 | 0.00 |
ENST00000381652.3
|
JAK2
|
Janus kinase 2 |
| chr12_-_75603643 | 0.00 |
ENST00000549446.1
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr8_+_97773202 | 0.00 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
Network of associatons between targets according to the STRING database.
First level regulatory network of PTF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.4 | 1.1 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.2 | 1.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 0.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 0.8 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.7 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.5 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.0 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 0.3 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.3 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.6 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.1 | 0.5 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 1.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.6 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 | 0.5 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.1 | 0.3 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0097021 | positive regulation of metanephric glomerulus development(GO:0072300) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 4.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.8 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0001654 | eye development(GO:0001654) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.4 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.1 | 1.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.4 | 4.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.3 | 1.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.0 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.6 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.1 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.3 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.1 | 0.5 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.7 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |