Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
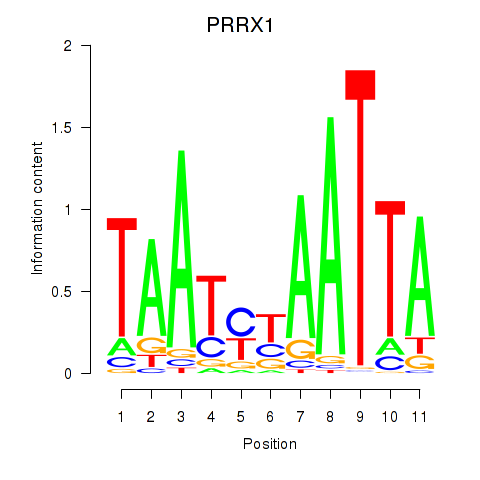
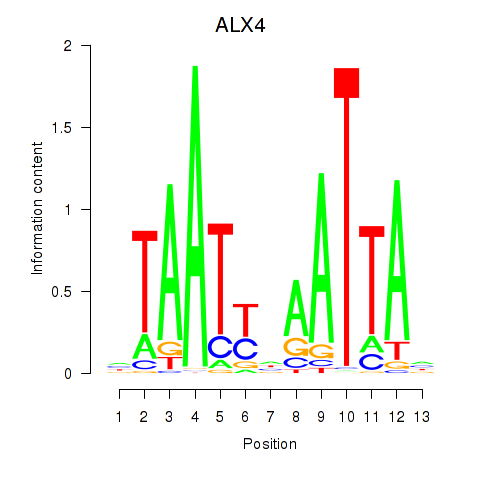
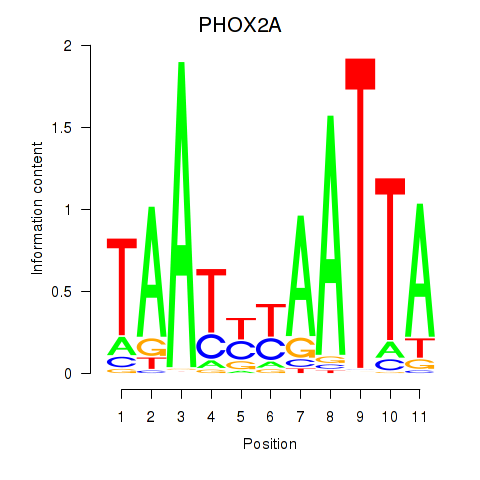
Results for PRRX1_ALX4_PHOX2A
Z-value: 0.55
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.7 | paired related homeobox 1 |
|
ALX4
|
ENSG00000052850.5 | ALX homeobox 4 |
|
PHOX2A
|
ENSG00000165462.5 | paired like homeobox 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ALX4 | hg19_v2_chr11_-_44331679_44331716 | -0.63 | 3.7e-01 | Click! |
| PRRX1 | hg19_v2_chr1_+_170633047_170633084 | -0.33 | 6.7e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_195310802 | 1.04 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr4_+_155484155 | 0.97 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr7_+_30589829 | 0.91 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr8_-_95229531 | 0.86 |
ENST00000450165.2
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr4_-_76649546 | 0.82 |
ENST00000508510.1
ENST00000509561.1 ENST00000499709.2 ENST00000511868.1 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_+_26087646 | 0.81 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr4_+_155484103 | 0.79 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr7_-_111032971 | 0.78 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr6_-_32908792 | 0.68 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_-_107235331 | 0.49 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr11_+_1295809 | 0.47 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr1_-_200379180 | 0.46 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr6_-_32908765 | 0.45 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr1_-_241799232 | 0.45 |
ENST00000366553.1
|
CHML
|
choroideremia-like (Rab escort protein 2) |
| chr3_-_27764190 | 0.45 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr12_+_14572070 | 0.44 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr1_-_151148492 | 0.44 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr18_+_20714525 | 0.41 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr2_-_114461655 | 0.41 |
ENST00000424612.1
|
AC017074.2
|
AC017074.2 |
| chr4_+_113568207 | 0.40 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr14_-_38064198 | 0.40 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chrX_+_77166172 | 0.37 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_+_53412368 | 0.37 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr6_-_107235378 | 0.35 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr6_-_107235287 | 0.35 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr3_-_87325612 | 0.34 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr2_+_44001172 | 0.33 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr14_+_38065052 | 0.30 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr3_+_149192475 | 0.30 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr14_-_83262540 | 0.30 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr15_-_76304731 | 0.29 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr3_+_149191723 | 0.29 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr1_-_234667504 | 0.28 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr7_-_55620433 | 0.27 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_+_183774240 | 0.26 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr16_+_53133070 | 0.26 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_17752912 | 0.25 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr3_-_186262166 | 0.24 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr8_-_17752996 | 0.24 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr8_-_102803163 | 0.24 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr1_-_151148442 | 0.24 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chr11_-_96076334 | 0.22 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr5_+_140514782 | 0.22 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr2_-_55496174 | 0.21 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr3_+_153839149 | 0.21 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr17_-_39156138 | 0.21 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr8_+_107738240 | 0.21 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr3_-_9291063 | 0.21 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr15_-_37393406 | 0.20 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr12_-_49581152 | 0.19 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_+_48485341 | 0.19 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr17_+_53342311 | 0.19 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr8_-_124741451 | 0.18 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr4_+_54927213 | 0.18 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr7_-_140482926 | 0.18 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr4_+_109571740 | 0.18 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr7_-_35013217 | 0.18 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr12_+_49621658 | 0.18 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr15_+_57511609 | 0.17 |
ENST00000543579.1
ENST00000537840.1 ENST00000343827.3 |
TCF12
|
transcription factor 12 |
| chr4_+_76649797 | 0.17 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chrX_+_119737806 | 0.17 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr12_-_118628350 | 0.17 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_136070614 | 0.17 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_+_50459990 | 0.16 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr3_-_130465604 | 0.16 |
ENST00000356763.3
|
PIK3R4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr6_+_3259122 | 0.16 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr5_-_134735568 | 0.16 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr12_-_91573132 | 0.15 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr2_+_32390925 | 0.15 |
ENST00000440718.1
ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr2_-_58468437 | 0.15 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr15_-_34331243 | 0.15 |
ENST00000306730.3
|
AVEN
|
apoptosis, caspase activation inhibitor |
| chr5_-_111093759 | 0.15 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr2_+_191334212 | 0.14 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr16_+_21244986 | 0.14 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr1_+_154401791 | 0.14 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr15_-_75748143 | 0.13 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_-_26231589 | 0.13 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chrX_+_149887090 | 0.13 |
ENST00000538506.1
|
MTMR1
|
myotubularin related protein 1 |
| chr19_+_50016610 | 0.13 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_+_76649753 | 0.13 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr9_+_42671887 | 0.13 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr1_+_81106951 | 0.13 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr9_+_124329336 | 0.13 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr1_-_242162375 | 0.13 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr14_-_57197224 | 0.13 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr1_+_62439037 | 0.12 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr6_+_3259148 | 0.12 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr12_-_118796910 | 0.12 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr5_-_70272055 | 0.12 |
ENST00000514857.2
|
NAIP
|
NLR family, apoptosis inhibitory protein |
| chr8_+_26150628 | 0.12 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr11_-_63439013 | 0.12 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr4_+_109571800 | 0.12 |
ENST00000512478.2
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr3_-_11623804 | 0.12 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr5_-_87516448 | 0.12 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr8_+_107738343 | 0.11 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr19_-_49149553 | 0.11 |
ENST00000084798.4
|
CA11
|
carbonic anhydrase XI |
| chr22_+_25615489 | 0.11 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr17_-_39023462 | 0.11 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr12_-_107380846 | 0.11 |
ENST00000548101.1
ENST00000550496.1 ENST00000552029.1 |
MTERFD3
|
MTERF domain containing 3 |
| chr12_-_118796971 | 0.11 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_200379129 | 0.11 |
ENST00000367353.1
|
ZNF281
|
zinc finger protein 281 |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr8_+_87111059 | 0.11 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr14_+_88851874 | 0.11 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr17_+_44701402 | 0.10 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr17_+_71228740 | 0.10 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr21_+_17442799 | 0.10 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrX_-_48858630 | 0.10 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr14_+_24099318 | 0.10 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr20_+_31823792 | 0.10 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr9_-_123639445 | 0.10 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr17_-_56494908 | 0.10 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr6_+_44187242 | 0.10 |
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_+_55980331 | 0.10 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr6_-_41039567 | 0.10 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr16_+_53469525 | 0.10 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr3_-_33759699 | 0.09 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr9_-_123639304 | 0.09 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr6_+_44187334 | 0.09 |
ENST00000313248.7
ENST00000427851.2 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr4_+_165878100 | 0.09 |
ENST00000513876.2
|
FAM218A
|
family with sequence similarity 218, member A |
| chr10_+_18689637 | 0.09 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_-_76072719 | 0.09 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_+_212475148 | 0.09 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr2_-_145275828 | 0.09 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_197300194 | 0.09 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr9_-_71869642 | 0.09 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr4_+_142558078 | 0.09 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr21_-_43346790 | 0.09 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chrX_-_48858667 | 0.08 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr11_-_92931098 | 0.08 |
ENST00000326402.4
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr3_-_32544900 | 0.08 |
ENST00000205636.3
|
CMTM6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr12_+_58003935 | 0.08 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr11_+_66276550 | 0.08 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr10_-_105110890 | 0.08 |
ENST00000369847.3
|
PCGF6
|
polycomb group ring finger 6 |
| chr10_-_75226166 | 0.08 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chrX_-_68385354 | 0.08 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr5_+_133562095 | 0.08 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chr1_+_170115142 | 0.08 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr10_-_105110831 | 0.08 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr12_-_107380910 | 0.08 |
ENST00000392830.2
ENST00000240050.4 |
MTERFD3
|
MTERF domain containing 3 |
| chr5_-_142780280 | 0.08 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr9_-_39239171 | 0.07 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr11_+_18433840 | 0.07 |
ENST00000541669.1
ENST00000280704.4 |
LDHC
|
lactate dehydrogenase C |
| chr12_+_116985896 | 0.07 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr14_-_21567009 | 0.07 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr7_+_138145076 | 0.07 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr6_+_88106840 | 0.07 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr9_+_2159850 | 0.07 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_52155001 | 0.07 |
ENST00000544199.1
|
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr9_-_123639600 | 0.07 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr12_+_20968608 | 0.07 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_-_16761117 | 0.07 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_136501478 | 0.07 |
ENST00000393056.2
ENST00000263611.2 |
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr5_+_68860949 | 0.07 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr17_-_79623597 | 0.07 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr11_-_111649015 | 0.07 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_+_151591422 | 0.06 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr7_-_27169801 | 0.06 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr17_+_71228537 | 0.06 |
ENST00000577615.1
ENST00000585109.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr12_+_72061563 | 0.06 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chrX_-_68385274 | 0.06 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr9_-_215744 | 0.06 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr12_-_118628315 | 0.06 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr15_+_75628394 | 0.06 |
ENST00000564815.1
ENST00000338995.6 |
COMMD4
|
COMM domain containing 4 |
| chr12_-_53601000 | 0.06 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr2_-_183291741 | 0.06 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr14_+_39734482 | 0.06 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr15_-_52970820 | 0.06 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr15_+_93426514 | 0.06 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chrX_+_9431324 | 0.06 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr10_+_67330096 | 0.06 |
ENST00000433152.4
ENST00000601979.1 ENST00000599409.1 ENST00000608678.1 |
RP11-222A11.1
|
RP11-222A11.1 |
| chr2_-_175711133 | 0.06 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr9_-_95244781 | 0.06 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr5_-_54603368 | 0.06 |
ENST00000508346.1
ENST00000251636.5 |
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr15_+_75628232 | 0.06 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chr8_-_80942061 | 0.06 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr2_+_223725652 | 0.05 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr3_+_140660634 | 0.05 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr9_-_95298314 | 0.05 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr1_+_186798073 | 0.05 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_-_13915530 | 0.05 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr1_+_145525015 | 0.05 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr1_+_180601139 | 0.05 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr12_+_100897130 | 0.05 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_+_36152163 | 0.05 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_-_15815626 | 0.05 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr14_+_21566980 | 0.05 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr11_-_82681626 | 0.05 |
ENST00000534396.1
|
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr17_-_5321549 | 0.05 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr5_-_111312622 | 0.05 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr13_-_26795840 | 0.05 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr10_+_89420706 | 0.05 |
ENST00000427144.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr11_+_43333513 | 0.05 |
ENST00000534695.1
ENST00000455725.2 ENST00000531273.1 ENST00000420461.2 ENST00000378852.3 ENST00000534600.1 |
API5
|
apoptosis inhibitor 5 |
| chr2_-_70529180 | 0.05 |
ENST00000450256.1
ENST00000037869.3 |
FAM136A
|
family with sequence similarity 136, member A |
| chr2_+_36923830 | 0.05 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr9_-_27005686 | 0.05 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chrX_+_48687283 | 0.04 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr2_+_170440844 | 0.04 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr4_-_83769996 | 0.04 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr1_-_209957882 | 0.04 |
ENST00000294811.1
|
C1orf74
|
chromosome 1 open reading frame 74 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:2001190 | peptide antigen assembly with MHC class II protein complex(GO:0002503) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.4 | 0.4 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.3 | 0.8 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.3 | 1.0 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 0.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 0.9 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 1.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.1 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.0 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 2.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.8 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.0 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.4 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 1.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 1.8 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |