Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
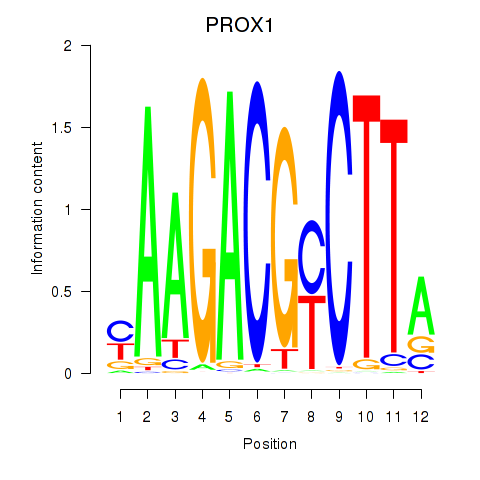
Results for PROX1
Z-value: 0.88
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322 | -0.98 | 2.4e-02 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_57308979 | 1.42 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr12_+_42624050 | 0.96 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr8_+_142264664 | 0.59 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr12_+_52668394 | 0.58 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr3_+_136676707 | 0.58 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr16_+_31724618 | 0.57 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr3_-_122283424 | 0.57 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr16_+_85832146 | 0.57 |
ENST00000565078.1
|
COX4I1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr22_-_39640756 | 0.51 |
ENST00000331163.6
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr21_-_16125773 | 0.50 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr19_-_45909585 | 0.50 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr5_-_157002775 | 0.49 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr2_-_233877912 | 0.49 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr6_+_90192974 | 0.46 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr7_+_48128316 | 0.45 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr7_+_48128194 | 0.45 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr20_-_61885826 | 0.45 |
ENST00000370316.3
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4 |
| chr19_-_48018203 | 0.44 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr2_-_163175133 | 0.44 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr1_-_109399682 | 0.43 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr8_+_145064233 | 0.43 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr8_+_145064215 | 0.41 |
ENST00000313269.5
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr18_-_2571210 | 0.39 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr8_-_19615538 | 0.39 |
ENST00000517494.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr5_-_157002749 | 0.38 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr3_-_48723268 | 0.38 |
ENST00000439518.1
ENST00000416649.2 ENST00000341520.4 ENST00000294129.2 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr19_-_40732594 | 0.37 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr18_-_21166841 | 0.37 |
ENST00000269228.5
|
NPC1
|
Niemann-Pick disease, type C1 |
| chr22_-_45559540 | 0.37 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr22_+_18121356 | 0.36 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr7_+_128864848 | 0.36 |
ENST00000325006.3
ENST00000446544.2 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr8_-_19615435 | 0.35 |
ENST00000523262.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr3_-_27410847 | 0.32 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr17_+_77018896 | 0.32 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr5_+_175490540 | 0.32 |
ENST00000515817.1
|
FAM153B
|
family with sequence similarity 153, member B |
| chr19_-_12267524 | 0.30 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr4_-_170947485 | 0.30 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr3_-_25915189 | 0.30 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr22_-_45559642 | 0.30 |
ENST00000426282.2
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr12_+_10331605 | 0.29 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr15_-_78913628 | 0.29 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr4_+_4861385 | 0.29 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr14_-_36645674 | 0.28 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr16_+_28857916 | 0.28 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr3_-_196065248 | 0.28 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr7_+_100209979 | 0.27 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr7_-_143582450 | 0.27 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr7_+_100210133 | 0.27 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr1_-_28241024 | 0.27 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr2_+_131862872 | 0.27 |
ENST00000439822.2
|
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr1_-_201476274 | 0.27 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr16_+_58059470 | 0.27 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr6_+_126102292 | 0.26 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_+_138067314 | 0.26 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr22_+_45559722 | 0.26 |
ENST00000347635.4
ENST00000407019.2 ENST00000424634.1 ENST00000417702.1 ENST00000425733.2 ENST00000430547.1 |
NUP50
|
nucleoporin 50kDa |
| chr1_-_201476220 | 0.25 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr9_+_132096166 | 0.24 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chr19_+_4007644 | 0.24 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr6_+_144665237 | 0.24 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr22_+_18121562 | 0.24 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr9_+_95858485 | 0.23 |
ENST00000375464.2
|
C9orf89
|
chromosome 9 open reading frame 89 |
| chr16_+_28985542 | 0.23 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr1_-_44497118 | 0.23 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_128886532 | 0.23 |
ENST00000444616.1
ENST00000388795.5 |
C4orf29
|
chromosome 4 open reading frame 29 |
| chr2_+_131862900 | 0.22 |
ENST00000438882.2
ENST00000538982.1 ENST00000404460.1 |
PLEKHB2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr2_+_108443388 | 0.22 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr7_+_100209725 | 0.22 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr12_+_54402790 | 0.22 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_-_28241226 | 0.22 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr5_+_179125368 | 0.22 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr1_+_171454639 | 0.22 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr17_+_72209653 | 0.22 |
ENST00000269346.4
|
TTYH2
|
tweety family member 2 |
| chr10_+_45869652 | 0.21 |
ENST00000542434.1
ENST00000374391.2 |
ALOX5
|
arachidonate 5-lipoxygenase |
| chr2_-_97534312 | 0.21 |
ENST00000442264.1
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr11_-_57298187 | 0.20 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr7_+_1609765 | 0.20 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr10_-_94301107 | 0.20 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr22_-_43539346 | 0.20 |
ENST00000327555.5
ENST00000290429.6 |
MCAT
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chrX_-_153236620 | 0.20 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr11_-_59383617 | 0.19 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr11_+_1874200 | 0.19 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
| chr2_+_241499719 | 0.19 |
ENST00000405954.1
|
DUSP28
|
dual specificity phosphatase 28 |
| chr6_-_72129806 | 0.19 |
ENST00000413945.1
ENST00000602878.1 ENST00000436803.1 ENST00000421704.1 ENST00000441570.1 |
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr6_-_134495992 | 0.19 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_-_46234119 | 0.19 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr4_-_69215699 | 0.19 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr16_-_22012419 | 0.19 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr8_-_19615382 | 0.19 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr10_+_71075552 | 0.19 |
ENST00000298649.3
|
HK1
|
hexokinase 1 |
| chr3_-_33138592 | 0.18 |
ENST00000415454.1
|
GLB1
|
galactosidase, beta 1 |
| chrX_-_153236819 | 0.18 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr11_-_73882029 | 0.18 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr2_+_28618532 | 0.18 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chrX_-_153237258 | 0.18 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr2_-_239140011 | 0.17 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr12_+_113229737 | 0.17 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr22_-_36761081 | 0.17 |
ENST00000456729.1
ENST00000401701.1 |
MYH9
|
myosin, heavy chain 9, non-muscle |
| chr19_-_16582815 | 0.17 |
ENST00000455140.2
ENST00000248070.6 ENST00000594975.1 |
EPS15L1
|
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr19_+_49535169 | 0.17 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr1_-_44497024 | 0.16 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_+_75136717 | 0.16 |
ENST00000297784.5
|
TMC1
|
transmembrane channel-like 1 |
| chrY_+_22918021 | 0.16 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr12_+_121078355 | 0.16 |
ENST00000316803.3
|
CABP1
|
calcium binding protein 1 |
| chr11_+_64008525 | 0.16 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr17_-_36904437 | 0.16 |
ENST00000585100.1
ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2
|
polycomb group ring finger 2 |
| chr2_-_65659762 | 0.16 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr14_+_78266408 | 0.16 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr21_-_16126181 | 0.16 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr14_+_73603126 | 0.16 |
ENST00000557356.1
ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1
|
presenilin 1 |
| chr8_-_41166953 | 0.15 |
ENST00000220772.3
|
SFRP1
|
secreted frizzled-related protein 1 |
| chr16_+_30669720 | 0.15 |
ENST00000356166.6
|
FBRS
|
fibrosin |
| chr11_-_111749767 | 0.15 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr1_+_95975672 | 0.15 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr6_+_34725263 | 0.15 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr19_-_53324884 | 0.15 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr10_-_52008313 | 0.15 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr16_-_12070468 | 0.15 |
ENST00000538896.1
|
RP11-166B2.1
|
Putative NPIP-like protein LOC729978 |
| chr2_-_239140276 | 0.15 |
ENST00000334973.4
|
AC016757.3
|
Protein LOC151174 |
| chr19_+_6361754 | 0.15 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr12_+_113229543 | 0.15 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr10_+_60028818 | 0.14 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr1_+_10459111 | 0.14 |
ENST00000541529.1
ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD
|
phosphogluconate dehydrogenase |
| chrX_-_53461305 | 0.14 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chrX_+_150565038 | 0.14 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr5_-_22853429 | 0.14 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr11_+_61722629 | 0.14 |
ENST00000526988.1
|
BEST1
|
bestrophin 1 |
| chr6_+_170151708 | 0.13 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr16_+_22518495 | 0.13 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr6_+_170151887 | 0.13 |
ENST00000588451.1
|
ERMARD
|
ER membrane-associated RNA degradation |
| chr4_+_128886424 | 0.13 |
ENST00000398965.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr3_-_137834436 | 0.12 |
ENST00000327532.2
ENST00000467030.1 |
DZIP1L
|
DAZ interacting zinc finger protein 1-like |
| chr8_+_117963190 | 0.12 |
ENST00000427715.2
|
SLC30A8
|
solute carrier family 30 (zinc transporter), member 8 |
| chr11_-_64739542 | 0.12 |
ENST00000536065.1
|
C11orf85
|
chromosome 11 open reading frame 85 |
| chr12_+_113229452 | 0.12 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr5_+_177433973 | 0.12 |
ENST00000507848.1
|
FAM153C
|
family with sequence similarity 153, member C |
| chr6_-_159420780 | 0.12 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr11_+_60609537 | 0.12 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr19_-_19754404 | 0.11 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chrX_+_49019061 | 0.11 |
ENST00000376339.1
ENST00000425661.2 ENST00000458388.1 ENST00000412696.2 |
MAGIX
|
MAGI family member, X-linked |
| chr22_-_32058166 | 0.11 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr9_-_130890662 | 0.11 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chr11_-_58035732 | 0.11 |
ENST00000395079.2
|
OR10W1
|
olfactory receptor, family 10, subfamily W, member 1 |
| chr4_-_159592996 | 0.11 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr17_+_80009330 | 0.11 |
ENST00000580716.1
ENST00000583961.1 |
GPS1
|
G protein pathway suppressor 1 |
| chr22_-_32058416 | 0.11 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_-_94050668 | 0.11 |
ENST00000539242.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr19_-_15529790 | 0.10 |
ENST00000596195.1
ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L
|
A kinase (PRKA) anchor protein 8-like |
| chr3_-_47555167 | 0.10 |
ENST00000296149.4
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr3_+_9839335 | 0.10 |
ENST00000453882.1
|
ARPC4-TTLL3
|
ARPC4-TTLL3 readthrough |
| chr8_-_116681686 | 0.10 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr3_-_100565249 | 0.10 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_-_112218378 | 0.10 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr9_-_130497565 | 0.10 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr9_-_127533582 | 0.10 |
ENST00000416460.2
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr6_-_137540477 | 0.10 |
ENST00000367735.2
ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chr17_+_36908984 | 0.10 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr16_+_23194033 | 0.10 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr18_+_32073253 | 0.10 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr11_-_61062762 | 0.10 |
ENST00000335613.5
|
VWCE
|
von Willebrand factor C and EGF domains |
| chr11_+_92085262 | 0.09 |
ENST00000298047.6
ENST00000409404.2 ENST00000541502.1 |
FAT3
|
FAT atypical cadherin 3 |
| chr18_+_60190226 | 0.09 |
ENST00000269499.5
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr12_-_122240792 | 0.09 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chrY_-_20935572 | 0.09 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr16_-_18801643 | 0.09 |
ENST00000322989.4
ENST00000563390.1 |
RPS15A
|
ribosomal protein S15a |
| chr5_-_177210399 | 0.09 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr10_+_104005272 | 0.09 |
ENST00000369983.3
|
GBF1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr14_-_54425475 | 0.09 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr22_+_26138108 | 0.09 |
ENST00000536101.1
ENST00000335473.7 ENST00000407587.2 |
MYO18B
|
myosin XVIIIB |
| chr6_-_42690312 | 0.09 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr9_+_125132803 | 0.09 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr10_-_50970322 | 0.09 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr1_-_15850676 | 0.09 |
ENST00000440484.1
ENST00000333868.5 |
CASP9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr15_-_58357932 | 0.08 |
ENST00000347587.3
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr22_+_24990746 | 0.08 |
ENST00000456869.1
ENST00000411974.1 |
GGT1
|
gamma-glutamyltransferase 1 |
| chr6_-_25042231 | 0.08 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr13_+_27131887 | 0.08 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr6_+_168196210 | 0.08 |
ENST00000597278.1
|
AL009178.1
|
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
| chr5_+_173473662 | 0.08 |
ENST00000519717.1
|
NSG2
|
Neuron-specific protein family member 2 |
| chr12_-_58240470 | 0.08 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr16_-_23607598 | 0.08 |
ENST00000562133.1
ENST00000570319.1 ENST00000007516.3 |
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
| chr15_-_90892669 | 0.08 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr1_+_161719552 | 0.08 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chrY_+_20708557 | 0.08 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr6_-_90529418 | 0.08 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr7_-_99716952 | 0.08 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr17_-_48546324 | 0.08 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr18_+_29769978 | 0.08 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr12_-_66524482 | 0.08 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr12_+_72079842 | 0.08 |
ENST00000266673.5
ENST00000550524.1 |
TMEM19
|
transmembrane protein 19 |
| chr11_-_46408107 | 0.08 |
ENST00000433765.2
|
CHRM4
|
cholinergic receptor, muscarinic 4 |
| chr22_-_29075853 | 0.08 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr19_+_6361440 | 0.07 |
ENST00000245816.4
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr15_-_75249793 | 0.07 |
ENST00000322177.5
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr16_-_28857677 | 0.07 |
ENST00000313511.3
|
TUFM
|
Tu translation elongation factor, mitochondrial |
| chr19_+_36120009 | 0.07 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr15_-_31453359 | 0.07 |
ENST00000542188.1
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr11_+_73359936 | 0.07 |
ENST00000542389.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_+_37321823 | 0.07 |
ENST00000487950.1
ENST00000469731.1 |
RNF8
|
ring finger protein 8, E3 ubiquitin protein ligase |
| chr9_-_127533519 | 0.07 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_-_4400415 | 0.07 |
ENST00000598564.1
ENST00000417295.2 ENST00000269886.3 |
SH3GL1
|
SH3-domain GRB2-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.1 | 0.6 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.6 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.4 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.5 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.9 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.4 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.2 | GO:2000053 | negative regulation of B cell differentiation(GO:0045578) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 1.4 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0071893 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:0045399 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) beta selection(GO:0043366) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) gamma-secretase complex(GO:0070765) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.9 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.4 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.6 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.0 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |