Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
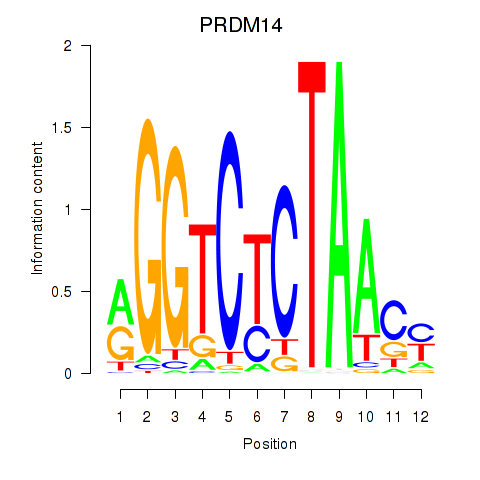
Results for PRDM14
Z-value: 0.49
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | 0.00 | 1.0e+00 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_34250861 | 0.57 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr14_+_21492331 | 0.34 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr3_-_134092561 | 0.34 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr15_+_63188009 | 0.31 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr9_+_132597722 | 0.25 |
ENST00000372429.3
ENST00000315480.4 ENST00000358355.1 |
USP20
|
ubiquitin specific peptidase 20 |
| chr11_-_65625014 | 0.22 |
ENST00000534784.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr1_+_155006300 | 0.20 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr5_-_55777586 | 0.19 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr1_+_39796810 | 0.16 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr17_+_75450075 | 0.16 |
ENST00000592951.1
|
SEPT9
|
septin 9 |
| chr12_-_124456598 | 0.15 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr1_+_27114589 | 0.15 |
ENST00000431541.1
ENST00000449950.2 ENST00000374145.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_-_155006254 | 0.14 |
ENST00000295536.5
|
DCST2
|
DC-STAMP domain containing 2 |
| chr1_+_36621529 | 0.14 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr2_-_208030295 | 0.14 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_49760639 | 0.14 |
ENST00000549538.1
ENST00000548654.1 ENST00000550643.1 ENST00000548710.1 ENST00000549179.1 ENST00000548377.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr19_+_49838653 | 0.14 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr19_-_6279932 | 0.14 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr5_+_32710736 | 0.13 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_+_36621174 | 0.13 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr19_+_39109735 | 0.13 |
ENST00000593149.1
ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K
|
eukaryotic translation initiation factor 3, subunit K |
| chr3_-_114477962 | 0.12 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_-_49351228 | 0.12 |
ENST00000541959.1
ENST00000447318.2 |
ARF3
|
ADP-ribosylation factor 3 |
| chr17_+_36873677 | 0.11 |
ENST00000471200.1
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr12_-_49351148 | 0.11 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr17_-_17485731 | 0.11 |
ENST00000395783.1
|
PEMT
|
phosphatidylethanolamine N-methyltransferase |
| chr11_-_46615498 | 0.10 |
ENST00000533727.1
ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1
|
autophagy/beclin-1 regulator 1 |
| chr15_+_45694567 | 0.10 |
ENST00000559860.1
|
SPATA5L1
|
spermatogenesis associated 5-like 1 |
| chr7_-_139763521 | 0.09 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr17_-_17726907 | 0.08 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr17_-_46692457 | 0.08 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr11_-_65624415 | 0.08 |
ENST00000524553.1
ENST00000527344.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr1_-_155006224 | 0.07 |
ENST00000368424.3
|
DCST2
|
DC-STAMP domain containing 2 |
| chr17_-_73775839 | 0.07 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr21_+_26934165 | 0.07 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr17_-_46691990 | 0.07 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr17_-_19651668 | 0.07 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr1_+_109102652 | 0.07 |
ENST00000370035.3
ENST00000405454.1 |
FAM102B
|
family with sequence similarity 102, member B |
| chr19_-_53757941 | 0.06 |
ENST00000594517.1
ENST00000601413.1 ENST00000594681.1 |
ZNF677
|
zinc finger protein 677 |
| chr17_-_19651654 | 0.06 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr12_-_49351303 | 0.06 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr3_-_47517302 | 0.06 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr6_+_43140095 | 0.05 |
ENST00000457278.2
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr2_-_192711968 | 0.05 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr1_+_27668505 | 0.05 |
ENST00000318074.5
|
SYTL1
|
synaptotagmin-like 1 |
| chr17_+_75450146 | 0.05 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr18_+_57567180 | 0.05 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chrX_+_51636629 | 0.05 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr12_+_121647962 | 0.05 |
ENST00000542067.1
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr16_-_55866997 | 0.04 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr12_+_121647868 | 0.04 |
ENST00000359949.7
ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr5_-_24645078 | 0.04 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr14_-_21492113 | 0.04 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr11_+_64879317 | 0.03 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr10_+_12171636 | 0.03 |
ENST00000379051.1
ENST00000379033.3 ENST00000441368.1 ENST00000298428.9 ENST00000304267.8 |
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr1_+_27114418 | 0.03 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chrX_+_41193407 | 0.03 |
ENST00000457138.2
ENST00000441189.2 |
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr3_+_10289707 | 0.03 |
ENST00000287652.4
|
TATDN2
|
TatD DNase domain containing 2 |
| chr17_+_75450099 | 0.03 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr11_+_85566422 | 0.03 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr6_+_133561725 | 0.02 |
ENST00000452339.2
|
EYA4
|
eyes absent homolog 4 (Drosophila) |
| chr1_+_153963227 | 0.02 |
ENST00000368567.4
ENST00000392558.4 |
RPS27
|
ribosomal protein S27 |
| chr12_-_56326402 | 0.02 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr6_-_109777128 | 0.01 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr7_+_39989611 | 0.01 |
ENST00000181839.4
|
CDK13
|
cyclin-dependent kinase 13 |
| chr15_+_69850521 | 0.01 |
ENST00000558781.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr19_+_3572758 | 0.00 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr17_-_2117600 | 0.00 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0051511 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |