Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
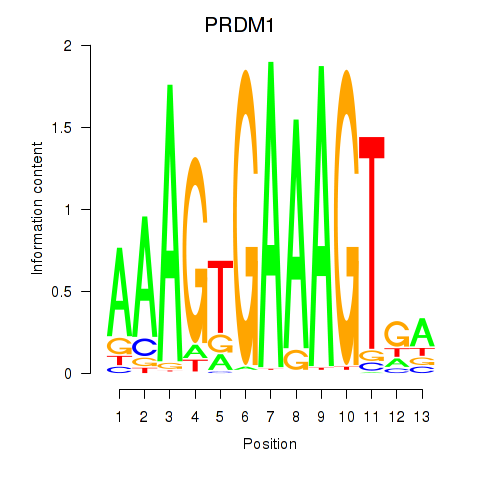
Results for PRDM1
Z-value: 0.43
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106546808_106546833 | -0.86 | 1.4e-01 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41158742 | 0.35 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr2_+_145780725 | 0.27 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr12_+_43086018 | 0.26 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr6_+_32821924 | 0.25 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_-_231084659 | 0.25 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr2_-_231084820 | 0.25 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr3_-_49851313 | 0.25 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr14_-_24615805 | 0.23 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_+_74372662 | 0.23 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr1_+_207262881 | 0.23 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr22_-_36635563 | 0.22 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr2_-_231084617 | 0.22 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr22_-_36635225 | 0.22 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr22_-_36635684 | 0.20 |
ENST00000358502.5
|
APOL2
|
apolipoprotein L, 2 |
| chr22_-_36556821 | 0.20 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr6_+_32811885 | 0.19 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr5_+_112227311 | 0.19 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr11_+_64323098 | 0.18 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_64323156 | 0.18 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr1_-_44497118 | 0.18 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr9_+_137533615 | 0.17 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr22_-_31688431 | 0.17 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_-_24615523 | 0.17 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr22_-_31688381 | 0.16 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_-_41742697 | 0.16 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr19_+_19516561 | 0.16 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr22_-_30642728 | 0.15 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr11_+_64323428 | 0.15 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_46402482 | 0.15 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_+_231191875 | 0.15 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr17_-_26694979 | 0.15 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr12_+_9144626 | 0.14 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr7_+_134832808 | 0.14 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr5_-_66942617 | 0.14 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr20_+_388679 | 0.14 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr11_+_46402744 | 0.14 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr12_-_120763739 | 0.14 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr12_+_6561190 | 0.14 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr10_-_82049424 | 0.13 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr8_+_39770803 | 0.13 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_+_64008525 | 0.13 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr21_+_43933946 | 0.13 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr12_+_94542459 | 0.13 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chr17_-_26695013 | 0.12 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr20_+_388935 | 0.12 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr20_+_388791 | 0.12 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_+_2985760 | 0.12 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr6_-_10413112 | 0.12 |
ENST00000465858.1
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr22_-_30642782 | 0.11 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chrX_-_118826784 | 0.11 |
ENST00000394616.4
|
SEPT6
|
septin 6 |
| chr22_-_36635598 | 0.11 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr11_+_46402583 | 0.10 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_71710648 | 0.10 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr11_+_64008443 | 0.10 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr8_+_77593448 | 0.10 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr6_+_32811861 | 0.10 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr16_-_72206034 | 0.10 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr17_-_56606664 | 0.10 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr8_-_79717163 | 0.10 |
ENST00000520269.1
|
IL7
|
interleukin 7 |
| chr17_-_56606705 | 0.09 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr6_+_26156551 | 0.09 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr18_+_18943554 | 0.09 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr11_+_46402297 | 0.09 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr5_+_73109339 | 0.09 |
ENST00000296799.4
|
ARHGEF28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr5_-_94417314 | 0.09 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_-_43339453 | 0.08 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr2_+_231280954 | 0.08 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr3_+_119013185 | 0.08 |
ENST00000264245.4
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr8_-_54436491 | 0.08 |
ENST00000426023.1
|
RP11-400K9.4
|
RP11-400K9.4 |
| chr6_-_32806483 | 0.08 |
ENST00000374899.4
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_+_134203273 | 0.08 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr17_-_43339474 | 0.08 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr10_-_131762105 | 0.07 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr6_-_32806506 | 0.07 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr13_+_24144796 | 0.07 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr5_-_94417186 | 0.07 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_+_110453109 | 0.07 |
ENST00000525659.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr11_+_71710973 | 0.07 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr2_+_179184955 | 0.07 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr13_-_33924755 | 0.07 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chrX_-_118827113 | 0.07 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr6_-_112575912 | 0.07 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr19_+_24009879 | 0.07 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr2_-_142888573 | 0.07 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr1_+_151372010 | 0.07 |
ENST00000290541.6
|
PSMB4
|
proteasome (prosome, macropain) subunit, beta type, 4 |
| chr16_-_88851618 | 0.07 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr11_+_71709938 | 0.06 |
ENST00000393705.4
ENST00000337131.5 ENST00000531053.1 ENST00000404792.1 |
IL18BP
|
interleukin 18 binding protein |
| chr19_-_17186229 | 0.06 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr22_+_39436862 | 0.06 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr4_-_111558135 | 0.06 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr2_+_145780767 | 0.06 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr1_-_44497024 | 0.06 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_-_32811771 | 0.06 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_-_167275991 | 0.06 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr18_+_34124507 | 0.06 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr17_-_56606639 | 0.06 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr6_+_53659746 | 0.06 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr9_+_18474163 | 0.05 |
ENST00000380566.4
|
ADAMTSL1
|
ADAMTS-like 1 |
| chr8_-_116681686 | 0.05 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr14_+_24605361 | 0.05 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr1_+_17906970 | 0.05 |
ENST00000375415.1
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_-_47621229 | 0.05 |
ENST00000434451.1
|
TNS3
|
tensin 3 |
| chr14_-_21490653 | 0.05 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chrX_-_118827333 | 0.05 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr12_-_49581152 | 0.05 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr5_-_94417562 | 0.05 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_+_7465216 | 0.05 |
ENST00000321337.7
|
SENP3
|
SUMO1/sentrin/SMT3 specific peptidase 3 |
| chr5_-_94417339 | 0.05 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr12_-_95941987 | 0.04 |
ENST00000537435.2
|
USP44
|
ubiquitin specific peptidase 44 |
| chr12_-_57505121 | 0.04 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr20_-_18774614 | 0.04 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr2_+_203499901 | 0.04 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr6_-_112575687 | 0.04 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr5_+_96212185 | 0.04 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr2_+_29336855 | 0.04 |
ENST00000404424.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_60609537 | 0.04 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr10_+_122610853 | 0.04 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr6_-_112575758 | 0.04 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr1_+_156737292 | 0.04 |
ENST00000271526.4
ENST00000353233.3 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr3_+_159706537 | 0.04 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr11_-_72432950 | 0.04 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr21_+_22370608 | 0.04 |
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr12_-_57504975 | 0.04 |
ENST00000553397.1
ENST00000556259.1 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr14_-_21490590 | 0.04 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chr5_-_64064508 | 0.04 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr12_-_106641728 | 0.03 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr14_+_24605389 | 0.03 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_+_234545148 | 0.03 |
ENST00000373445.1
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr13_+_102104980 | 0.03 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_-_168864427 | 0.03 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr4_-_46996424 | 0.03 |
ENST00000264318.3
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chr15_+_85144217 | 0.03 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr7_-_3083573 | 0.03 |
ENST00000396946.4
|
CARD11
|
caspase recruitment domain family, member 11 |
| chr3_+_107241783 | 0.03 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_99797522 | 0.03 |
ENST00000389677.5
|
FAXC
|
failed axon connections homolog (Drosophila) |
| chr8_-_23540402 | 0.03 |
ENST00000523261.1
ENST00000380871.4 |
NKX3-1
|
NK3 homeobox 1 |
| chr9_+_18474098 | 0.03 |
ENST00000327883.7
ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1
|
ADAMTS-like 1 |
| chr16_-_67970990 | 0.03 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr3_+_47422485 | 0.03 |
ENST00000431726.1
ENST00000456221.1 ENST00000265562.4 |
PTPN23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr8_-_29120604 | 0.03 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr12_+_10163231 | 0.03 |
ENST00000396502.1
ENST00000338896.5 |
CLEC12B
|
C-type lectin domain family 12, member B |
| chr8_-_29120580 | 0.03 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr2_+_231280908 | 0.03 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr20_+_6748311 | 0.02 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr19_+_17186577 | 0.02 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr14_-_21490417 | 0.02 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr6_-_167276033 | 0.02 |
ENST00000503859.1
ENST00000506565.1 |
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr10_+_91589261 | 0.02 |
ENST00000448963.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr17_+_6659153 | 0.02 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr11_-_85338311 | 0.02 |
ENST00000376104.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr7_+_95401851 | 0.02 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr3_+_8543561 | 0.02 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr12_-_52800139 | 0.02 |
ENST00000257974.2
|
KRT82
|
keratin 82 |
| chr18_-_47017956 | 0.02 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr8_-_116680833 | 0.02 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr22_+_41697520 | 0.02 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr10_+_91589309 | 0.02 |
ENST00000448490.1
|
LINC00865
|
long intergenic non-protein coding RNA 865 |
| chr1_-_153919128 | 0.02 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr4_+_146403912 | 0.02 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr15_-_72565340 | 0.02 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr17_-_79818354 | 0.02 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr13_+_24144509 | 0.02 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr16_+_31085714 | 0.02 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr4_-_140544386 | 0.02 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr15_+_75491213 | 0.02 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_+_2985726 | 0.02 |
ENST00000511072.1
ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16
|
PR domain containing 16 |
| chr1_-_161014731 | 0.02 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr17_+_3379284 | 0.02 |
ENST00000263080.2
|
ASPA
|
aspartoacylase |
| chr5_-_59481406 | 0.02 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_199996702 | 0.02 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr20_+_13976015 | 0.02 |
ENST00000217246.4
|
MACROD2
|
MACRO domain containing 2 |
| chr11_+_128563948 | 0.01 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_110453203 | 0.01 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr2_-_145275228 | 0.01 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_81203972 | 0.01 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr6_-_29527702 | 0.01 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr8_-_116681221 | 0.01 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr8_+_77318769 | 0.01 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr11_+_128563652 | 0.01 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_35104185 | 0.01 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr6_-_41909191 | 0.01 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr8_+_77593474 | 0.01 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr1_-_111746966 | 0.01 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr4_+_152020736 | 0.01 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr1_+_164600184 | 0.01 |
ENST00000482110.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_152020715 | 0.01 |
ENST00000274065.4
|
RPS3A
|
ribosomal protein S3A |
| chr9_+_133454943 | 0.01 |
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3 |
| chr19_+_13106383 | 0.01 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr18_-_53070913 | 0.01 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr6_-_138820624 | 0.01 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr16_-_3930724 | 0.01 |
ENST00000262367.5
|
CREBBP
|
CREB binding protein |
| chr20_+_30946106 | 0.01 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr6_-_154831779 | 0.01 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr7_-_143059845 | 0.01 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chr2_+_234590556 | 0.01 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr12_-_118498911 | 0.01 |
ENST00000544233.1
|
WSB2
|
WD repeat and SOCS box containing 2 |
| chr12_-_50561075 | 0.01 |
ENST00000422340.2
ENST00000317551.6 |
CERS5
|
ceramide synthase 5 |
| chr7_+_128355439 | 0.00 |
ENST00000315184.5
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr8_-_56986768 | 0.00 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr13_+_108922228 | 0.00 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0097227 | sperm annulus(GO:0097227) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.7 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |