Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
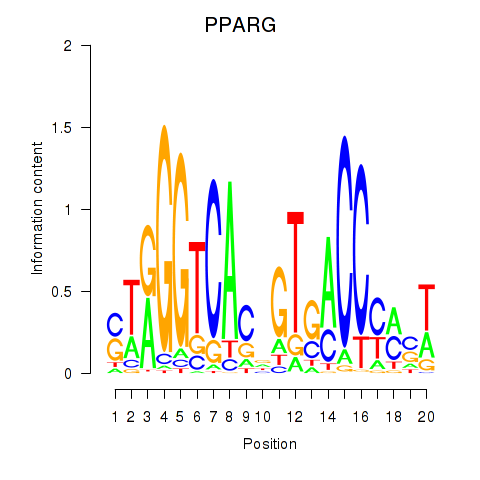
Results for PPARG
Z-value: 1.01
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | peroxisome proliferator activated receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12329358_12329393 | -0.63 | 3.7e-01 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_111970353 | 0.36 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr2_+_112895939 | 0.34 |
ENST00000331203.2
ENST00000409903.1 ENST00000409667.3 ENST00000409450.3 |
FBLN7
|
fibulin 7 |
| chr3_+_238969 | 0.32 |
ENST00000421198.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr19_-_55740632 | 0.29 |
ENST00000327042.4
|
TMEM86B
|
transmembrane protein 86B |
| chr19_+_544034 | 0.26 |
ENST00000592501.1
ENST00000264553.3 |
GZMM
|
granzyme M (lymphocyte met-ase 1) |
| chr3_-_123710893 | 0.24 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr7_+_75932863 | 0.22 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr19_-_39303576 | 0.21 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr10_+_97667360 | 0.20 |
ENST00000602648.1
|
C10orf131
|
chromosome 10 open reading frame 131 |
| chr19_-_22018966 | 0.19 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr19_+_49661079 | 0.18 |
ENST00000355712.5
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr17_-_48450534 | 0.18 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr16_+_84209738 | 0.18 |
ENST00000564928.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr19_-_1592828 | 0.18 |
ENST00000592012.1
|
MBD3
|
methyl-CpG binding domain protein 3 |
| chr21_+_38338737 | 0.18 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr19_-_51587502 | 0.18 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chr22_+_50312316 | 0.17 |
ENST00000328268.4
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr8_-_145642203 | 0.17 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr19_-_55836669 | 0.17 |
ENST00000326652.4
|
TMEM150B
|
transmembrane protein 150B |
| chr7_-_99569468 | 0.17 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr14_+_100848311 | 0.17 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr11_+_65687158 | 0.17 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr11_+_65686952 | 0.16 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_13068012 | 0.16 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr9_-_38424443 | 0.16 |
ENST00000377694.1
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1 |
| chr1_-_11115877 | 0.16 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chrX_-_106959631 | 0.16 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr1_-_1891537 | 0.15 |
ENST00000493316.1
ENST00000461752.2 |
C1orf222
|
chromosome 1 open reading frame 222 |
| chr14_+_105953204 | 0.15 |
ENST00000409393.2
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr6_+_41196052 | 0.15 |
ENST00000341495.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr11_+_47587129 | 0.15 |
ENST00000326656.8
ENST00000326674.9 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr13_-_52026730 | 0.15 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr12_-_80084594 | 0.14 |
ENST00000548426.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr10_+_97667722 | 0.14 |
ENST00000423344.2
|
C10orf131
|
chromosome 10 open reading frame 131 |
| chr17_-_74287333 | 0.14 |
ENST00000447564.2
|
QRICH2
|
glutamine rich 2 |
| chr12_+_57849048 | 0.14 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr14_+_105953246 | 0.14 |
ENST00000392531.3
|
CRIP1
|
cysteine-rich protein 1 (intestinal) |
| chr17_-_18945798 | 0.14 |
ENST00000395635.1
|
GRAP
|
GRB2-related adaptor protein |
| chr9_+_101984577 | 0.14 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr19_+_49661037 | 0.13 |
ENST00000427978.2
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr16_-_85969774 | 0.13 |
ENST00000598933.1
|
RP11-542M13.3
|
RP11-542M13.3 |
| chr17_-_48450265 | 0.13 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr19_-_55836697 | 0.13 |
ENST00000438693.1
ENST00000591570.1 |
TMEM150B
|
transmembrane protein 150B |
| chr19_+_4343691 | 0.13 |
ENST00000597036.1
|
MPND
|
MPN domain containing |
| chr19_+_58193388 | 0.13 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr21_+_45705752 | 0.13 |
ENST00000291582.5
|
AIRE
|
autoimmune regulator |
| chr19_+_44645700 | 0.13 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr12_-_56123444 | 0.13 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr14_+_105212297 | 0.12 |
ENST00000556623.1
ENST00000555674.1 |
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr1_-_220101944 | 0.12 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr22_-_21579843 | 0.12 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr22_+_50312274 | 0.12 |
ENST00000404488.3
|
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr11_-_2924720 | 0.12 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr8_-_145701718 | 0.11 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr1_-_47407136 | 0.11 |
ENST00000462347.1
ENST00000371905.1 ENST00000310638.4 |
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr19_-_10445399 | 0.11 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr17_+_41177220 | 0.11 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr12_-_56122761 | 0.11 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr18_-_5238525 | 0.11 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr19_-_6502304 | 0.11 |
ENST00000540257.1
ENST00000594276.1 ENST00000594075.1 ENST00000600216.1 ENST00000596926.1 |
TUBB4A
|
tubulin, beta 4A class IVa |
| chr22_+_22681656 | 0.11 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr22_+_24105241 | 0.11 |
ENST00000402217.3
|
C22orf15
|
chromosome 22 open reading frame 15 |
| chr15_+_40674963 | 0.11 |
ENST00000448395.2
|
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr16_+_50730910 | 0.11 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr4_+_87856191 | 0.11 |
ENST00000503477.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr2_+_220306745 | 0.11 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr7_-_38407770 | 0.11 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr4_-_682769 | 0.11 |
ENST00000507165.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr18_+_12947126 | 0.11 |
ENST00000592170.1
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr17_+_48450575 | 0.10 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr8_+_125551338 | 0.10 |
ENST00000276689.3
ENST00000518008.1 ENST00000522532.1 ENST00000517367.1 |
NDUFB9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa |
| chr22_+_47070490 | 0.10 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr19_-_16653226 | 0.10 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr1_-_223853425 | 0.10 |
ENST00000366873.2
ENST00000419193.2 |
CAPN8
|
calpain 8 |
| chr3_+_125687987 | 0.10 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr19_+_4472230 | 0.10 |
ENST00000301284.4
ENST00000586684.1 |
HDGFRP2
|
Hepatoma-derived growth factor-related protein 2 |
| chrX_+_153770421 | 0.10 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr6_-_33160231 | 0.10 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr8_-_42752418 | 0.10 |
ENST00000524954.1
|
RNF170
|
ring finger protein 170 |
| chr19_-_16653325 | 0.10 |
ENST00000546361.2
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr21_-_33984456 | 0.10 |
ENST00000431216.1
ENST00000553001.1 ENST00000440966.1 |
AP000275.65
C21orf59
|
Uncharacterized protein chromosome 21 open reading frame 59 |
| chr6_+_31783291 | 0.10 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr19_+_1438383 | 0.10 |
ENST00000586686.2
ENST00000591032.1 ENST00000586656.1 ENST00000589656.2 ENST00000593052.1 ENST00000586096.2 ENST00000591804.2 |
RPS15
|
ribosomal protein S15 |
| chr9_+_139847347 | 0.10 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr19_+_18682531 | 0.09 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr9_-_130616915 | 0.09 |
ENST00000344849.3
|
ENG
|
endoglin |
| chr11_+_65686802 | 0.09 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr22_+_50312379 | 0.09 |
ENST00000407217.3
ENST00000403427.3 |
CRELD2
|
cysteine-rich with EGF-like domains 2 |
| chr6_-_33168391 | 0.09 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr16_+_90089008 | 0.09 |
ENST00000268699.4
|
GAS8
|
growth arrest-specific 8 |
| chr20_+_62152077 | 0.09 |
ENST00000370179.3
ENST00000370177.1 |
PPDPF
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr19_+_52800410 | 0.09 |
ENST00000595962.1
ENST00000598016.1 ENST00000334564.7 ENST00000490272.1 |
ZNF480
|
zinc finger protein 480 |
| chr12_+_112451120 | 0.09 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chrX_-_51812268 | 0.09 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr1_-_95392635 | 0.09 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr11_-_64901978 | 0.09 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr11_+_65686728 | 0.09 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_5680231 | 0.09 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr3_+_183873240 | 0.09 |
ENST00000431765.1
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr19_-_1592652 | 0.09 |
ENST00000156825.1
ENST00000434436.3 |
MBD3
|
methyl-CpG binding domain protein 3 |
| chr6_-_49712147 | 0.09 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr19_+_49660997 | 0.09 |
ENST00000598691.1
ENST00000252826.5 |
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr19_+_18682661 | 0.09 |
ENST00000596273.1
ENST00000442744.2 ENST00000595683.1 ENST00000599256.1 ENST00000595158.1 ENST00000598780.1 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr8_-_82024290 | 0.09 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr17_+_27071002 | 0.09 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr6_-_31628512 | 0.09 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr8_-_143696833 | 0.09 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr19_-_47349395 | 0.09 |
ENST00000597020.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr11_+_68816356 | 0.08 |
ENST00000294309.3
ENST00000542467.1 |
TPCN2
|
two pore segment channel 2 |
| chr11_+_46740730 | 0.08 |
ENST00000311907.5
ENST00000530231.1 ENST00000442468.1 |
F2
|
coagulation factor II (thrombin) |
| chrX_-_153979315 | 0.08 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr20_+_37434329 | 0.08 |
ENST00000299824.1
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B |
| chr9_-_139094988 | 0.08 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr20_+_49411523 | 0.08 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr12_+_109569155 | 0.08 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr18_+_55711575 | 0.08 |
ENST00000356462.6
ENST00000400345.3 ENST00000589054.1 ENST00000256832.7 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr21_+_47063590 | 0.08 |
ENST00000400314.1
|
PCBP3
|
poly(rC) binding protein 3 |
| chr10_-_99052382 | 0.08 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr19_+_18283959 | 0.08 |
ENST00000597802.2
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chr19_-_55919087 | 0.08 |
ENST00000587845.1
ENST00000589978.1 ENST00000264552.9 |
UBE2S
|
ubiquitin-conjugating enzyme E2S |
| chr22_+_19929130 | 0.08 |
ENST00000361682.6
ENST00000403184.1 ENST00000403710.1 ENST00000407537.1 |
COMT
|
catechol-O-methyltransferase |
| chr15_+_40675132 | 0.08 |
ENST00000608100.1
ENST00000557920.1 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr2_-_74779744 | 0.08 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr16_-_67969888 | 0.08 |
ENST00000574576.2
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr7_-_86688990 | 0.08 |
ENST00000450689.2
|
KIAA1324L
|
KIAA1324-like |
| chr2_-_74619152 | 0.08 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr19_-_2783363 | 0.08 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr13_+_48611665 | 0.08 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr22_+_29702996 | 0.08 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr1_-_109203997 | 0.08 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr19_-_46405861 | 0.08 |
ENST00000322217.5
|
MYPOP
|
Myb-related transcription factor, partner of profilin |
| chr1_+_203274639 | 0.08 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr6_-_30080876 | 0.08 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr8_-_143484483 | 0.08 |
ENST00000519651.1
ENST00000307180.3 ENST00000518720.1 ENST00000524325.1 |
TSNARE1
|
t-SNARE domain containing 1 |
| chr16_-_4850471 | 0.08 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr11_-_64901900 | 0.07 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr22_-_24641027 | 0.07 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr11_+_7506837 | 0.07 |
ENST00000528758.1
|
OLFML1
|
olfactomedin-like 1 |
| chr17_-_8093471 | 0.07 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr11_+_2923499 | 0.07 |
ENST00000449793.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr22_-_23484246 | 0.07 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr11_+_117070904 | 0.07 |
ENST00000529792.1
|
TAGLN
|
transgelin |
| chr7_-_99527243 | 0.07 |
ENST00000312891.2
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa |
| chr19_+_50016610 | 0.07 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chrX_+_153775821 | 0.07 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr11_+_2923619 | 0.07 |
ENST00000380574.1
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr12_-_57352103 | 0.07 |
ENST00000398138.3
|
RDH16
|
retinol dehydrogenase 16 (all-trans) |
| chr20_-_35402123 | 0.07 |
ENST00000373740.3
ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1
|
DSN1, MIS12 kinetochore complex component |
| chr17_-_57784755 | 0.07 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr4_+_57371509 | 0.07 |
ENST00000360096.2
|
ARL9
|
ADP-ribosylation factor-like 9 |
| chr10_+_74870253 | 0.07 |
ENST00000544879.1
ENST00000537969.1 ENST00000372997.3 |
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13 |
| chr10_-_48390973 | 0.07 |
ENST00000224600.4
|
RBP3
|
retinol binding protein 3, interstitial |
| chr19_+_17970677 | 0.07 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr19_+_42388437 | 0.07 |
ENST00000378152.4
ENST00000337665.4 |
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr17_-_79196799 | 0.07 |
ENST00000269392.4
|
AZI1
|
5-azacytidine induced 1 |
| chr18_+_5238549 | 0.07 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr16_+_770975 | 0.07 |
ENST00000569529.1
ENST00000564000.1 ENST00000219535.3 |
FAM173A
|
family with sequence similarity 173, member A |
| chrX_+_51928002 | 0.07 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr8_+_145729465 | 0.07 |
ENST00000394955.2
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase) |
| chr22_+_23229960 | 0.07 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr2_-_74618964 | 0.07 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr11_-_7904464 | 0.07 |
ENST00000502624.2
ENST00000527565.1 |
RP11-35J10.5
|
RP11-35J10.5 |
| chr16_+_476379 | 0.07 |
ENST00000434585.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr11_+_65292538 | 0.07 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr19_+_4343584 | 0.07 |
ENST00000596722.1
|
MPND
|
MPN domain containing |
| chr11_-_67210930 | 0.07 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
| chr16_-_15736345 | 0.06 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr3_+_51705844 | 0.06 |
ENST00000457927.1
ENST00000444233.1 |
TEX264
|
testis expressed 264 |
| chr21_-_33985127 | 0.06 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr8_-_107782463 | 0.06 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr4_-_860950 | 0.06 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr10_-_129924468 | 0.06 |
ENST00000368653.3
|
MKI67
|
marker of proliferation Ki-67 |
| chr8_-_145641864 | 0.06 |
ENST00000276833.5
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr11_-_9025541 | 0.06 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr19_-_5680202 | 0.06 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr5_-_179050066 | 0.06 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr17_-_77023723 | 0.06 |
ENST00000577521.1
|
C1QTNF1-AS1
|
C1QTNF1 antisense RNA 1 |
| chr17_+_55173933 | 0.06 |
ENST00000539273.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr19_-_45735138 | 0.06 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr1_+_220701456 | 0.06 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr5_+_176731572 | 0.06 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr11_+_60609537 | 0.06 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr22_-_50689786 | 0.06 |
ENST00000216271.5
|
HDAC10
|
histone deacetylase 10 |
| chr19_+_35609380 | 0.06 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3 |
| chr17_-_27045405 | 0.06 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr9_+_95709733 | 0.06 |
ENST00000375482.3
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr19_+_35629702 | 0.06 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr16_+_67022633 | 0.06 |
ENST00000398354.1
ENST00000326686.5 |
CES4A
|
carboxylesterase 4A |
| chr6_+_31795506 | 0.06 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr19_+_1491144 | 0.06 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chr19_-_45826125 | 0.06 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr1_+_20512568 | 0.06 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr17_-_74236382 | 0.06 |
ENST00000592271.1
ENST00000319945.6 ENST00000269391.6 |
RNF157
|
ring finger protein 157 |
| chr1_-_115323245 | 0.06 |
ENST00000060969.5
ENST00000369528.5 |
SIKE1
|
suppressor of IKBKE 1 |
| chr3_-_46786245 | 0.06 |
ENST00000442359.2
|
PRSS45
|
protease, serine, 45 |
| chr19_-_14228541 | 0.06 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr10_-_72362515 | 0.06 |
ENST00000373209.2
ENST00000441259.1 |
PRF1
|
perforin 1 (pore forming protein) |
| chr11_+_47586982 | 0.06 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr8_+_145203548 | 0.06 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr9_+_34458851 | 0.06 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000360 | negative regulation of fertilization(GO:0060467) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.4 | GO:1990736 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.2 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0060585 | activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0050666 | cellular response to phosphate starvation(GO:0016036) regulation of sulfur amino acid metabolic process(GO:0031335) positive regulation of sulfur amino acid metabolic process(GO:0031337) regulation of homocysteine metabolic process(GO:0050666) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.0 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.2 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.0 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.1 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0060589 | GTPase regulator activity(GO:0030695) nucleoside-triphosphatase regulator activity(GO:0060589) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |