Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
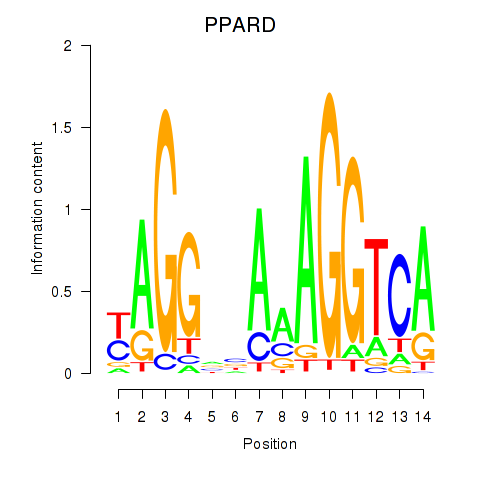
Results for PPARD
Z-value: 0.13
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARD | hg19_v2_chr6_+_35310391_35310410 | 0.95 | 4.5e-02 | Click! |
Activity profile of PPARD motif
Sorted Z-values of PPARD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_34978980 | 0.27 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr1_+_9005917 | 0.09 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr9_+_131037623 | 0.07 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr12_-_6665200 | 0.06 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr8_-_80942467 | 0.06 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr19_+_29493486 | 0.06 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr2_-_198364552 | 0.05 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr1_-_160253998 | 0.05 |
ENST00000392220.2
|
PEX19
|
peroxisomal biogenesis factor 19 |
| chr18_-_47340297 | 0.05 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr19_-_51869592 | 0.05 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr14_+_74551650 | 0.05 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr1_+_6511651 | 0.05 |
ENST00000434576.1
|
ESPN
|
espin |
| chr1_-_161039647 | 0.05 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr13_+_113760098 | 0.04 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr11_-_119599794 | 0.04 |
ENST00000264025.3
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chrX_-_108868390 | 0.04 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr1_-_161039753 | 0.04 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr1_+_145727681 | 0.04 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr19_-_10687948 | 0.04 |
ENST00000592285.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr2_+_228190066 | 0.04 |
ENST00000436237.1
ENST00000443428.2 ENST00000418961.1 |
MFF
|
mitochondrial fission factor |
| chr8_-_80942061 | 0.04 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr11_-_45939565 | 0.04 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_+_37894179 | 0.04 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr16_-_75284758 | 0.04 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr11_+_123325106 | 0.03 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr3_-_48936272 | 0.03 |
ENST00000544097.1
ENST00000430379.1 ENST00000319017.4 |
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr9_+_131901661 | 0.03 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr22_-_29784519 | 0.03 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr2_-_198364581 | 0.03 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr3_-_125094093 | 0.03 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr19_-_10687907 | 0.03 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr1_-_161039456 | 0.03 |
ENST00000368016.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr11_+_64073699 | 0.03 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr16_-_28303360 | 0.03 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr2_+_95940186 | 0.03 |
ENST00000403131.2
ENST00000317668.4 ENST00000317620.9 |
PROM2
|
prominin 2 |
| chr8_-_80942139 | 0.03 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr5_-_143550159 | 0.02 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr19_-_10687983 | 0.02 |
ENST00000587069.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_45939374 | 0.02 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr1_+_62417957 | 0.02 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr12_-_21757774 | 0.02 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr20_-_44485835 | 0.02 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr19_+_29493456 | 0.02 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr12_-_53320245 | 0.02 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr15_+_78441663 | 0.02 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr1_-_197036364 | 0.02 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr16_-_4401284 | 0.02 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr2_-_159313214 | 0.02 |
ENST00000409889.1
ENST00000283233.5 ENST00000536771.1 |
CCDC148
|
coiled-coil domain containing 148 |
| chr2_-_28113217 | 0.02 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr6_+_33172407 | 0.02 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chrX_+_2746818 | 0.02 |
ENST00000398806.3
|
GYG2
|
glycogenin 2 |
| chr9_-_33402506 | 0.02 |
ENST00000377425.4
ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7
|
aquaporin 7 |
| chr6_-_33547975 | 0.02 |
ENST00000442998.2
ENST00000360661.5 |
BAK1
|
BCL2-antagonist/killer 1 |
| chr2_+_73441350 | 0.02 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr12_-_109219937 | 0.02 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr1_+_165864821 | 0.02 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_165864800 | 0.01 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr3_-_50605077 | 0.01 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr1_+_43855545 | 0.01 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr4_-_7069760 | 0.01 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr10_-_94050820 | 0.01 |
ENST00000265997.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr14_+_32546274 | 0.01 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_-_207024134 | 0.01 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_-_109968973 | 0.01 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr1_+_202317855 | 0.01 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr6_-_33548006 | 0.01 |
ENST00000374467.3
|
BAK1
|
BCL2-antagonist/killer 1 |
| chr6_+_30295036 | 0.01 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr20_+_19870167 | 0.01 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr1_-_161102367 | 0.01 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr9_+_126773880 | 0.01 |
ENST00000373615.4
|
LHX2
|
LIM homeobox 2 |
| chr5_-_176981417 | 0.01 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr20_+_39969519 | 0.01 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr15_-_82555000 | 0.01 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr2_+_219433281 | 0.01 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr1_-_11863171 | 0.01 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr16_-_4401258 | 0.01 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr17_+_4853442 | 0.01 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr20_+_42187682 | 0.01 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chrX_+_2746850 | 0.01 |
ENST00000381163.3
ENST00000338623.5 ENST00000542787.1 |
GYG2
|
glycogenin 2 |
| chr2_+_178257372 | 0.01 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr1_-_43855479 | 0.01 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr3_+_19189946 | 0.01 |
ENST00000328405.2
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr22_-_51016846 | 0.01 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr14_-_51561784 | 0.01 |
ENST00000360392.4
|
TRIM9
|
tripartite motif containing 9 |
| chr20_+_32077880 | 0.01 |
ENST00000342704.6
ENST00000375279.2 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr7_-_87342564 | 0.01 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr11_-_116968987 | 0.01 |
ENST00000434315.2
ENST00000292055.4 ENST00000375288.1 ENST00000542607.1 ENST00000445177.1 ENST00000375300.1 ENST00000446921.2 |
SIK3
|
SIK family kinase 3 |
| chr5_+_14664762 | 0.01 |
ENST00000284274.4
|
FAM105B
|
family with sequence similarity 105, member B |
| chr17_+_48610074 | 0.01 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr2_+_219283815 | 0.01 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr4_+_174089904 | 0.01 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr15_+_43885252 | 0.00 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr16_+_2255710 | 0.00 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr20_+_42187608 | 0.00 |
ENST00000373100.1
|
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr12_+_96252706 | 0.00 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_+_159593418 | 0.00 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr16_+_2255841 | 0.00 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr15_+_75494214 | 0.00 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_-_29508321 | 0.00 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr2_+_120125245 | 0.00 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr1_-_120311517 | 0.00 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr11_+_111169565 | 0.00 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr11_+_66624527 | 0.00 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_+_81272287 | 0.00 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr17_+_73997419 | 0.00 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr9_+_35042205 | 0.00 |
ENST00000312292.5
ENST00000378745.3 |
C9orf131
|
chromosome 9 open reading frame 131 |
| chr10_-_49459800 | 0.00 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr16_-_74808710 | 0.00 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr4_+_159593271 | 0.00 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr17_+_7123125 | 0.00 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.0 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.0 | GO:0002352 | B cell selection(GO:0002339) B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |