Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
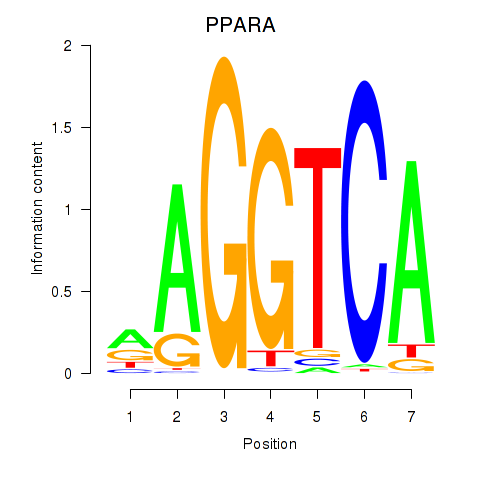
Results for PPARA
Z-value: 0.63
Transcription factors associated with PPARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARA
|
ENSG00000186951.12 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARA | hg19_v2_chr22_+_46546494_46546525 | -0.23 | 7.7e-01 | Click! |
Activity profile of PPARA motif
Sorted Z-values of PPARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_21492331 | 0.61 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr22_+_22676808 | 0.59 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr12_-_124873357 | 0.50 |
ENST00000448614.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr10_-_10504285 | 0.35 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr12_-_6665200 | 0.28 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr22_+_47158578 | 0.27 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr17_+_7533439 | 0.27 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr18_-_43678241 | 0.27 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr10_+_81065975 | 0.27 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chrX_-_21776281 | 0.26 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr17_-_42988004 | 0.26 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr15_+_75335604 | 0.25 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_-_117716418 | 0.23 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr22_-_39239987 | 0.22 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr22_+_37959647 | 0.22 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr12_+_93963590 | 0.21 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr4_-_40632140 | 0.21 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr1_-_16532985 | 0.20 |
ENST00000441785.1
ENST00000449495.1 |
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr2_-_231860596 | 0.20 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr16_+_3704822 | 0.20 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr20_+_43343886 | 0.19 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr19_-_51289374 | 0.19 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr17_-_30470154 | 0.19 |
ENST00000398832.2
|
AC090616.2
|
Uncharacterized protein |
| chr10_+_77056181 | 0.19 |
ENST00000526759.1
ENST00000533822.1 |
ZNF503-AS1
|
ZNF503 antisense RNA 1 |
| chrX_+_18725758 | 0.19 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr5_+_133451254 | 0.19 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr3_+_188817891 | 0.19 |
ENST00000412373.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr17_-_1553346 | 0.19 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr6_-_149969871 | 0.19 |
ENST00000335643.8
ENST00000444282.1 |
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr5_+_172332220 | 0.18 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr5_-_139930713 | 0.18 |
ENST00000602657.1
|
SRA1
|
steroid receptor RNA activator 1 |
| chr14_+_75894391 | 0.18 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr18_-_12377001 | 0.18 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr8_+_94929273 | 0.17 |
ENST00000518573.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr22_+_51176624 | 0.17 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr16_-_30997533 | 0.17 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr19_-_36019123 | 0.17 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_+_153600869 | 0.17 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr22_+_39101728 | 0.17 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr1_+_45140400 | 0.17 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr19_-_55881741 | 0.16 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr6_+_30749649 | 0.16 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr11_+_3877412 | 0.16 |
ENST00000527651.1
|
STIM1
|
stromal interaction molecule 1 |
| chr22_+_32149927 | 0.16 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr12_-_25348007 | 0.16 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr11_+_77899842 | 0.16 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr7_-_20826504 | 0.16 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr6_-_149969829 | 0.16 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr15_-_44487408 | 0.16 |
ENST00000402883.1
ENST00000417257.1 |
FRMD5
|
FERM domain containing 5 |
| chr2_-_207023918 | 0.16 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr2_+_65663812 | 0.15 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr19_-_12912601 | 0.15 |
ENST00000334482.5
|
PRDX2
|
peroxiredoxin 2 |
| chr19_-_49371711 | 0.15 |
ENST00000355496.5
ENST00000263265.6 |
PLEKHA4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr17_+_74536115 | 0.15 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr9_-_35685452 | 0.15 |
ENST00000607559.1
|
TPM2
|
tropomyosin 2 (beta) |
| chr2_-_202316169 | 0.15 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr22_+_23237555 | 0.15 |
ENST00000390321.2
|
IGLC1
|
immunoglobulin lambda constant 1 (Mcg marker) |
| chr8_+_86376081 | 0.15 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr20_+_19870167 | 0.15 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr6_-_34664612 | 0.15 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr15_-_72767490 | 0.15 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr12_+_56511943 | 0.14 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr17_+_7483761 | 0.14 |
ENST00000584180.1
|
CD68
|
CD68 molecule |
| chr16_+_69139467 | 0.14 |
ENST00000569188.1
|
HAS3
|
hyaluronan synthase 3 |
| chr19_+_35629702 | 0.14 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr15_-_44486632 | 0.14 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr5_+_135383008 | 0.14 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr5_-_149682447 | 0.14 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr17_+_7517264 | 0.14 |
ENST00000593717.1
ENST00000572182.1 ENST00000574539.1 ENST00000576728.1 ENST00000575314.1 ENST00000570547.1 ENST00000572262.1 ENST00000576478.1 |
AC007421.1
SHBG
|
AC007421.1 sex hormone-binding globulin |
| chr11_+_57308979 | 0.14 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr17_+_40440481 | 0.14 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr14_-_69444957 | 0.14 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr20_+_58179582 | 0.14 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr20_+_44462749 | 0.14 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr12_-_15865844 | 0.14 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr20_+_44486246 | 0.13 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr2_-_207024134 | 0.13 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr8_-_42698292 | 0.13 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr21_+_43823983 | 0.13 |
ENST00000291535.6
ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr8_+_96037255 | 0.13 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr3_+_14444063 | 0.13 |
ENST00000454876.2
ENST00000360861.3 ENST00000416216.2 |
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr10_+_99185917 | 0.13 |
ENST00000334828.5
|
PGAM1
|
phosphoglycerate mutase 1 (brain) |
| chr9_+_21440440 | 0.13 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr6_+_13272904 | 0.13 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr13_-_41635512 | 0.13 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr11_+_57365150 | 0.13 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr3_-_110612323 | 0.13 |
ENST00000383686.2
|
RP11-553A10.1
|
Uncharacterized protein |
| chr19_+_54466179 | 0.13 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr8_+_97597148 | 0.13 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr15_-_78423567 | 0.13 |
ENST00000561190.1
ENST00000559645.1 ENST00000560618.1 ENST00000559054.1 |
CIB2
|
calcium and integrin binding family member 2 |
| chr22_+_47158518 | 0.13 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr9_+_132388566 | 0.12 |
ENST00000372480.1
|
NTMT1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chrX_+_44732757 | 0.12 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr9_-_16705069 | 0.12 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr6_+_36839616 | 0.12 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr14_+_61654271 | 0.12 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr14_+_73525265 | 0.12 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr14_-_31889737 | 0.12 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr11_+_73498973 | 0.12 |
ENST00000537007.1
|
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr3_+_63638372 | 0.12 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr12_+_109577202 | 0.12 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr17_-_41174367 | 0.12 |
ENST00000587173.1
|
VAT1
|
vesicle amine transport 1 |
| chr12_-_46121554 | 0.12 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr11_-_45928830 | 0.12 |
ENST00000449465.1
|
C11orf94
|
chromosome 11 open reading frame 94 |
| chr11_-_34379546 | 0.12 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr7_+_35756186 | 0.12 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr8_-_107782463 | 0.12 |
ENST00000311955.3
|
ABRA
|
actin-binding Rho activating protein |
| chr17_-_70417365 | 0.12 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr1_+_206858328 | 0.12 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr12_+_54378849 | 0.12 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr4_-_65275162 | 0.12 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr1_-_33815486 | 0.12 |
ENST00000373418.3
|
PHC2
|
polyhomeotic homolog 2 (Drosophila) |
| chr6_-_2751146 | 0.11 |
ENST00000268446.5
ENST00000274643.7 |
MYLK4
|
myosin light chain kinase family, member 4 |
| chr19_+_41699135 | 0.11 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr11_+_111957497 | 0.11 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chrX_-_152989798 | 0.11 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr11_-_1783633 | 0.11 |
ENST00000367196.3
|
CTSD
|
cathepsin D |
| chr5_+_218356 | 0.11 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr8_+_110656344 | 0.11 |
ENST00000499579.1
|
RP11-422N16.3
|
Uncharacterized protein |
| chr2_-_198364552 | 0.11 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr2_+_228678550 | 0.11 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr17_+_79670386 | 0.11 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr20_+_35807714 | 0.11 |
ENST00000373632.4
|
RPN2
|
ribophorin II |
| chr17_+_2699697 | 0.11 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr3_-_110612059 | 0.11 |
ENST00000485473.1
|
RP11-553A10.1
|
Uncharacterized protein |
| chr6_+_31939608 | 0.11 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr3_-_124839648 | 0.11 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr7_-_105029812 | 0.11 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr22_+_21321447 | 0.11 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr17_-_74722536 | 0.11 |
ENST00000585429.1
|
JMJD6
|
jumonji domain containing 6 |
| chr12_-_96184913 | 0.11 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr19_+_49109990 | 0.11 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr7_+_129007964 | 0.11 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr12_-_109219937 | 0.11 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr22_-_29784519 | 0.10 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_+_28874860 | 0.10 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr12_-_16760195 | 0.10 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_103574024 | 0.10 |
ENST00000512756.1
ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1
|
collagen, type XI, alpha 1 |
| chr1_+_199996733 | 0.10 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr11_+_57435441 | 0.10 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr6_-_31107127 | 0.10 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr2_+_232457569 | 0.10 |
ENST00000313965.2
|
C2orf57
|
chromosome 2 open reading frame 57 |
| chr8_-_145550337 | 0.10 |
ENST00000531896.1
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr19_+_10197463 | 0.10 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr17_-_39216344 | 0.10 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr6_+_149068464 | 0.10 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr20_+_45947246 | 0.10 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr10_+_104178946 | 0.10 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chrX_-_108868390 | 0.10 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr6_-_36953833 | 0.10 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr11_+_77899920 | 0.10 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr8_-_94928861 | 0.10 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2 |
| chrX_+_47050798 | 0.10 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr15_+_92397051 | 0.10 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr12_+_54426637 | 0.10 |
ENST00000312492.2
|
HOXC5
|
homeobox C5 |
| chr1_-_150693318 | 0.10 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr1_-_19536744 | 0.10 |
ENST00000375267.2
ENST00000375217.2 ENST00000375226.2 ENST00000375254.3 |
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr1_-_229569834 | 0.10 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr17_+_74536164 | 0.10 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr1_-_201140673 | 0.10 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr5_-_173217931 | 0.09 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr2_+_152214098 | 0.09 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr6_+_36973406 | 0.09 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr3_-_50340996 | 0.09 |
ENST00000266031.4
ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1
|
hyaluronoglucosaminidase 1 |
| chr1_+_19923454 | 0.09 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr1_+_169077133 | 0.09 |
ENST00000494797.1
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr1_-_35325318 | 0.09 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
| chr17_-_79900255 | 0.09 |
ENST00000330655.3
ENST00000582198.1 |
MYADML2
PYCR1
|
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr17_+_13972807 | 0.09 |
ENST00000429152.2
ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10
|
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr9_-_16728161 | 0.09 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr14_-_23395623 | 0.09 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr12_-_48214345 | 0.09 |
ENST00000433685.1
ENST00000447463.1 ENST00000427332.2 |
HDAC7
|
histone deacetylase 7 |
| chr5_+_179125907 | 0.09 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr14_+_75894714 | 0.09 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr1_-_154164534 | 0.09 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr5_+_129240151 | 0.09 |
ENST00000305031.4
|
CHSY3
|
chondroitin sulfate synthase 3 |
| chr17_+_19281787 | 0.09 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr2_+_134877740 | 0.09 |
ENST00000409645.1
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr11_-_66103932 | 0.09 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr8_-_53626974 | 0.09 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr15_+_67418047 | 0.09 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr17_+_73975292 | 0.09 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr17_-_74722672 | 0.09 |
ENST00000397625.4
ENST00000445478.2 |
JMJD6
|
jumonji domain containing 6 |
| chr20_+_3776371 | 0.09 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr1_-_157014865 | 0.09 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr22_+_30792980 | 0.09 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr12_+_6833323 | 0.09 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr16_-_74734742 | 0.09 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr7_+_45613958 | 0.09 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr3_-_141747950 | 0.09 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr22_-_38699003 | 0.09 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chrX_+_70521584 | 0.09 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr4_+_108852697 | 0.09 |
ENST00000508453.1
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1 |
| chr12_+_6976687 | 0.09 |
ENST00000396705.5
|
TPI1
|
triosephosphate isomerase 1 |
| chr1_+_155179012 | 0.09 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr1_-_150849047 | 0.09 |
ENST00000354396.2
ENST00000505755.1 |
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr10_-_127464390 | 0.09 |
ENST00000368808.3
|
MMP21
|
matrix metallopeptidase 21 |
| chr5_+_154181816 | 0.09 |
ENST00000518677.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr15_+_65337708 | 0.09 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr9_+_115983808 | 0.09 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.1 | GO:0035568 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0002339 | B cell selection(GO:0002339) B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0045362 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:1903278 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.0 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.3 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.4 | GO:0070470 | plasma membrane respiratory chain complex I(GO:0045272) plasma membrane respiratory chain(GO:0070470) |
| 0.0 | 0.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:1990175 | EH domain binding(GO:1990175) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.0 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |