Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
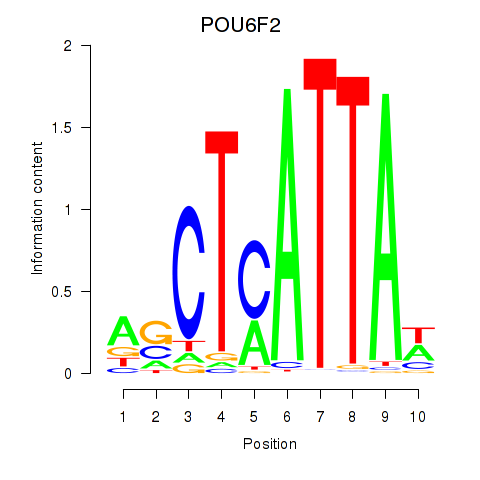
Results for POU6F2
Z-value: 0.55
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU class 6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg19_v2_chr7_+_39125365_39125489 | -0.34 | 6.6e-01 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_53133070 | 0.53 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_3469181 | 0.51 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chrX_+_43515467 | 0.50 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr15_+_96869165 | 0.45 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_37393406 | 0.41 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr17_+_48243352 | 0.40 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr4_+_66536248 | 0.37 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr11_-_27494279 | 0.37 |
ENST00000379214.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_-_27494309 | 0.37 |
ENST00000389858.4
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr17_+_73539339 | 0.34 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr18_-_24129367 | 0.34 |
ENST00000408011.3
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr1_+_61869748 | 0.32 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr15_+_58702742 | 0.31 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chrX_+_10031499 | 0.30 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr7_-_16840820 | 0.30 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr13_+_98612446 | 0.29 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr12_+_28410128 | 0.28 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr2_+_102721023 | 0.27 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr17_-_27418537 | 0.26 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr2_+_138721850 | 0.25 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr11_+_30344595 | 0.25 |
ENST00000282032.3
|
ARL14EP
|
ADP-ribosylation factor-like 14 effector protein |
| chr19_+_9203855 | 0.25 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr13_-_110438914 | 0.24 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr10_-_92681033 | 0.23 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr16_-_54962704 | 0.22 |
ENST00000502066.2
ENST00000560912.1 ENST00000558952.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr1_-_193028426 | 0.22 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr2_+_44502597 | 0.22 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr15_-_54025300 | 0.22 |
ENST00000559418.1
|
WDR72
|
WD repeat domain 72 |
| chr6_-_33160231 | 0.22 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr3_-_127541194 | 0.22 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr6_-_111136513 | 0.22 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr11_-_31531121 | 0.21 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chrX_+_86772707 | 0.20 |
ENST00000373119.4
|
KLHL4
|
kelch-like family member 4 |
| chr1_+_193028717 | 0.19 |
ENST00000415442.2
ENST00000506303.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr17_-_39165366 | 0.19 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr16_-_54962415 | 0.19 |
ENST00000501177.3
ENST00000559598.2 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr17_+_4613918 | 0.19 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr10_+_94352956 | 0.19 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr1_-_204329013 | 0.18 |
ENST00000272203.3
ENST00000414478.1 |
PLEKHA6
|
pleckstrin homology domain containing, family A member 6 |
| chr19_-_47137942 | 0.18 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr4_+_175204818 | 0.18 |
ENST00000503780.1
|
CEP44
|
centrosomal protein 44kDa |
| chr15_+_93443419 | 0.17 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr2_+_44502630 | 0.17 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr21_+_17442799 | 0.17 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_-_216003127 | 0.16 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr1_+_193028552 | 0.16 |
ENST00000400968.2
ENST00000432079.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr13_-_44735393 | 0.16 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr20_+_58515417 | 0.16 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr11_-_129062093 | 0.16 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr7_-_55620433 | 0.16 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr15_+_75080883 | 0.16 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr5_+_115177178 | 0.15 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_+_19536083 | 0.15 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr1_-_193028621 | 0.15 |
ENST00000367455.4
ENST00000367454.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chrX_-_38186775 | 0.15 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr15_-_33447055 | 0.15 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr12_-_86230315 | 0.14 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr6_-_32157947 | 0.14 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr20_+_5987890 | 0.14 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr9_+_131902283 | 0.13 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_-_159832438 | 0.13 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr11_-_63439013 | 0.13 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr7_+_115862858 | 0.13 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr21_+_17443521 | 0.13 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_4613776 | 0.13 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr15_+_101402041 | 0.13 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr3_+_141106458 | 0.12 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr15_+_96904487 | 0.12 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr3_+_189507432 | 0.12 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr5_-_54988448 | 0.11 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr8_-_42396185 | 0.11 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr6_-_111804905 | 0.11 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr11_-_93583697 | 0.11 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr6_+_130339710 | 0.11 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_+_73539232 | 0.11 |
ENST00000580925.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr8_+_26150628 | 0.11 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr8_-_142012169 | 0.10 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chrX_+_86772787 | 0.10 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr3_+_189507460 | 0.10 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr8_+_22424551 | 0.10 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_-_106697974 | 0.09 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr7_-_83824449 | 0.09 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr19_-_7698599 | 0.09 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr21_+_17792672 | 0.09 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_-_11610255 | 0.09 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr17_-_46716647 | 0.09 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chrX_-_38186811 | 0.09 |
ENST00000318842.7
|
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr20_+_56136136 | 0.08 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr4_+_175204865 | 0.08 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr8_+_42396274 | 0.08 |
ENST00000438528.3
|
SMIM19
|
small integral membrane protein 19 |
| chr17_-_2117600 | 0.08 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr11_+_31531291 | 0.08 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr3_-_57199397 | 0.07 |
ENST00000296318.7
|
IL17RD
|
interleukin 17 receptor D |
| chr1_-_193028632 | 0.07 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr1_-_234667504 | 0.07 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr7_+_116660246 | 0.07 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr20_-_58515344 | 0.07 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chrX_-_151619746 | 0.06 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr11_-_71823715 | 0.06 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr19_+_50016610 | 0.06 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_-_109541539 | 0.06 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr3_+_189507523 | 0.06 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr17_-_33390667 | 0.06 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr13_+_46039037 | 0.06 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr11_-_71823266 | 0.06 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr13_+_78315466 | 0.06 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_-_30877837 | 0.05 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_-_94257512 | 0.05 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr12_-_15815626 | 0.05 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr17_-_29624343 | 0.05 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr6_-_74231444 | 0.05 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_71823796 | 0.05 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr5_-_111093167 | 0.05 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr1_+_200011711 | 0.05 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_16759440 | 0.04 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chrX_-_19988382 | 0.04 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr3_+_147657764 | 0.04 |
ENST00000467198.1
ENST00000485006.1 |
RP11-71N10.1
|
RP11-71N10.1 |
| chr5_+_137722255 | 0.04 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr14_+_101359265 | 0.04 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr20_+_42187682 | 0.04 |
ENST00000373092.3
ENST00000373077.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr5_+_136070614 | 0.04 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr11_-_36619771 | 0.04 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr8_+_77593474 | 0.04 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr10_+_114710211 | 0.04 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_+_159343688 | 0.04 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr17_+_18086392 | 0.04 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr22_+_20905286 | 0.04 |
ENST00000428629.1
|
MED15
|
mediator complex subunit 15 |
| chr21_-_35899113 | 0.03 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr11_+_7598239 | 0.03 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr13_-_74993252 | 0.03 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr17_-_19290483 | 0.03 |
ENST00000395592.2
ENST00000299610.4 |
MFAP4
|
microfibrillar-associated protein 4 |
| chr2_+_128403439 | 0.03 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr14_+_32798547 | 0.03 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr12_-_16761117 | 0.03 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_+_4087664 | 0.03 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr2_+_128403720 | 0.03 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr14_-_27066960 | 0.02 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr5_+_67586465 | 0.02 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_74486109 | 0.02 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_175204765 | 0.02 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr12_-_71031185 | 0.02 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr6_-_8064567 | 0.02 |
ENST00000543936.1
ENST00000397457.2 |
BLOC1S5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr14_-_50698276 | 0.02 |
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila) |
| chr1_+_10271674 | 0.02 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr11_-_82444892 | 0.01 |
ENST00000329203.3
|
FAM181B
|
family with sequence similarity 181, member B |
| chr4_+_86525299 | 0.01 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_-_16758873 | 0.01 |
ENST00000535535.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chrX_-_21676442 | 0.01 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr17_-_40337470 | 0.01 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr18_-_13915530 | 0.01 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr12_+_54332535 | 0.01 |
ENST00000243056.3
|
HOXC13
|
homeobox C13 |
| chr3_-_33686743 | 0.01 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr16_+_31885079 | 0.01 |
ENST00000300870.10
ENST00000394846.3 |
ZNF267
|
zinc finger protein 267 |
| chr2_-_70780572 | 0.01 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr11_+_45918092 | 0.01 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr3_+_130648842 | 0.01 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_+_59100774 | 0.01 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr1_+_46379254 | 0.01 |
ENST00000372008.2
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr17_+_37784749 | 0.00 |
ENST00000394265.1
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr6_-_111136299 | 0.00 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr7_+_141478242 | 0.00 |
ENST00000247881.2
|
TAS2R4
|
taste receptor, type 2, member 4 |
| chr1_+_62439037 | 0.00 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr7_-_73038822 | 0.00 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chr1_-_11024258 | 0.00 |
ENST00000418570.2
|
C1orf127
|
chromosome 1 open reading frame 127 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.3 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.4 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.3 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.3 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.2 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0030169 | triglyceride lipase activity(GO:0004806) low-density lipoprotein particle binding(GO:0030169) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |