Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
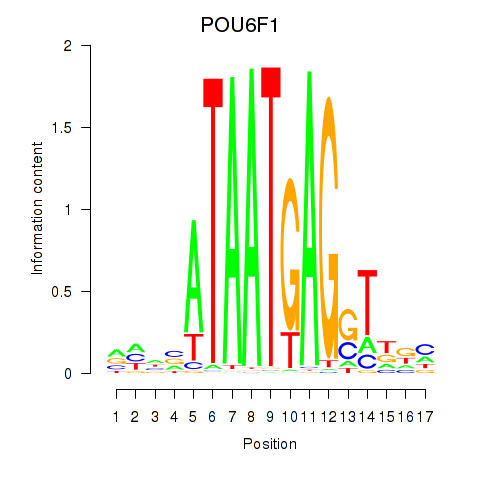
Results for POU6F1
Z-value: 0.45
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.11 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F1 | hg19_v2_chr12_-_51611477_51611612 | 0.33 | 6.7e-01 | Click! |
Activity profile of POU6F1 motif
Sorted Z-values of POU6F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_102513950 | 0.53 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr2_+_102721023 | 0.49 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr13_+_98612446 | 0.43 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr5_+_162864575 | 0.39 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr1_-_197115818 | 0.35 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr12_+_49297899 | 0.34 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chrX_+_43515467 | 0.34 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr13_-_52027134 | 0.25 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr9_+_26746951 | 0.21 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr1_+_225117350 | 0.21 |
ENST00000413949.2
ENST00000430092.1 ENST00000366850.3 ENST00000400952.3 ENST00000366849.1 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr8_-_146176199 | 0.21 |
ENST00000532351.1
ENST00000276816.4 ENST00000394909.2 |
ZNF16
|
zinc finger protein 16 |
| chr15_+_59279851 | 0.20 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr17_-_25568687 | 0.20 |
ENST00000581944.1
|
RP11-663N22.1
|
RP11-663N22.1 |
| chr11_+_85339623 | 0.19 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr19_+_10959043 | 0.19 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr20_+_58515417 | 0.19 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr2_+_242289502 | 0.19 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr7_-_83824449 | 0.19 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr18_+_52495426 | 0.18 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr1_+_225140372 | 0.18 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr17_+_73539339 | 0.17 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr20_+_5987890 | 0.17 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr5_+_115177178 | 0.16 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr6_-_47009996 | 0.16 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr12_-_57443886 | 0.15 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr15_-_85259360 | 0.15 |
ENST00000559729.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr22_+_31518938 | 0.15 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr12_+_102514019 | 0.15 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chr15_-_44828838 | 0.15 |
ENST00000560750.1
|
EIF3J-AS1
|
EIF3J antisense RNA 1 (head to head) |
| chr14_-_70883708 | 0.15 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chr10_+_97803151 | 0.14 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chrY_+_14958970 | 0.13 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr1_-_243326612 | 0.13 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr7_+_141490017 | 0.13 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr12_+_49297887 | 0.12 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr15_-_85259384 | 0.11 |
ENST00000455959.3
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr21_-_34915123 | 0.11 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr16_-_58718638 | 0.11 |
ENST00000562397.1
ENST00000564010.1 ENST00000570214.1 ENST00000563196.1 |
SLC38A7
|
solute carrier family 38, member 7 |
| chr2_-_85828867 | 0.10 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr1_-_161008697 | 0.09 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr3_-_48659193 | 0.09 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr6_-_111136513 | 0.09 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr6_-_111804905 | 0.09 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_-_100129128 | 0.08 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr11_-_93583697 | 0.08 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr1_-_151431909 | 0.08 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr7_-_77427676 | 0.08 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr2_+_191045562 | 0.08 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr15_-_31283618 | 0.08 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr15_-_85259294 | 0.07 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr15_-_85259330 | 0.07 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chrX_+_102024075 | 0.06 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr1_-_151431647 | 0.06 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr20_-_58515344 | 0.06 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chrX_-_122756660 | 0.06 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr14_+_24970800 | 0.06 |
ENST00000555109.1
|
RP11-80A15.1
|
Uncharacterized protein |
| chr11_-_65381643 | 0.06 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr12_-_7596735 | 0.05 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr6_-_26235206 | 0.05 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr18_+_9475001 | 0.05 |
ENST00000019317.4
|
RALBP1
|
ralA binding protein 1 |
| chr9_+_128509624 | 0.05 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr7_+_48494660 | 0.05 |
ENST00000411975.1
ENST00000544596.1 |
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr6_-_138539627 | 0.05 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr10_-_94257512 | 0.05 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr5_+_67586465 | 0.05 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr10_-_43904235 | 0.05 |
ENST00000356053.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr6_-_110501126 | 0.05 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chrX_+_119737806 | 0.04 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr11_+_118398178 | 0.04 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr16_+_19422035 | 0.04 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chr1_+_176432298 | 0.04 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr6_-_111136299 | 0.04 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr15_+_45028719 | 0.04 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr6_-_110501200 | 0.03 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr1_-_153538292 | 0.03 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chr17_-_39165366 | 0.03 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr17_-_40337470 | 0.02 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr9_+_103790991 | 0.02 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr2_+_128403720 | 0.02 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr4_-_83812157 | 0.02 |
ENST00000513323.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr1_-_151431674 | 0.02 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr2_+_128403439 | 0.02 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr20_-_50418972 | 0.02 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr4_+_40201954 | 0.01 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr12_-_95397442 | 0.01 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr18_-_73967160 | 0.01 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr19_+_42817527 | 0.01 |
ENST00000598766.1
|
TMEM145
|
transmembrane protein 145 |
| chr7_-_38407770 | 0.01 |
ENST00000390348.2
|
TRGV1
|
T cell receptor gamma variable 1 (non-functional) |
| chr22_-_28490123 | 0.01 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr9_+_131644398 | 0.01 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr2_-_179516225 | 0.01 |
ENST00000446966.1
ENST00000426232.1 |
TTN
|
titin |
| chr1_+_222913009 | 0.00 |
ENST00000456298.1
|
FAM177B
|
family with sequence similarity 177, member B |
| chr6_+_110501344 | 0.00 |
ENST00000368932.1
|
CDC40
|
cell division cycle 40 |
| chr9_-_4666337 | 0.00 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |