Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
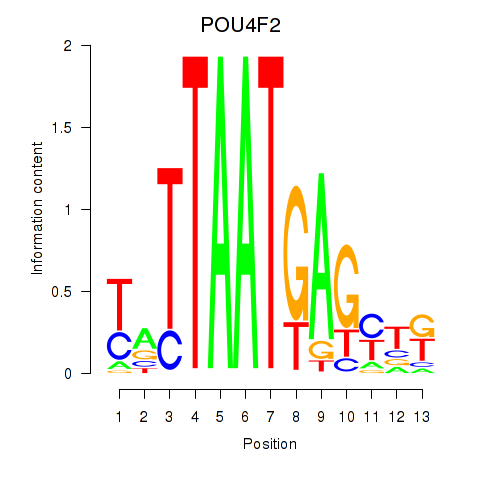
Results for POU4F2
Z-value: 0.18
Transcription factors associated with POU4F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F2
|
ENSG00000151615.3 | POU class 4 homeobox 2 |
Activity profile of POU4F2 motif
Sorted Z-values of POU4F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_134914673 | 0.28 |
ENST00000512158.1
|
CXCL14
|
chemokine (C-X-C motif) ligand 14 |
| chr4_-_169401628 | 0.18 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr19_-_38344165 | 0.14 |
ENST00000592714.1
ENST00000587395.1 |
AC016582.2
|
AC016582.2 |
| chr4_+_169013666 | 0.09 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr5_+_102200948 | 0.08 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_-_89746264 | 0.08 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chrX_-_106146547 | 0.07 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr11_-_107729287 | 0.06 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chrX_-_99987088 | 0.06 |
ENST00000372981.1
ENST00000263033.5 |
SYTL4
|
synaptotagmin-like 4 |
| chr9_-_5830768 | 0.05 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr11_-_107729504 | 0.05 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chrX_+_133930798 | 0.04 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr1_-_193028632 | 0.04 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr14_+_55221541 | 0.04 |
ENST00000555192.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr1_+_40506392 | 0.04 |
ENST00000414893.1
ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr11_-_107729887 | 0.04 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_89746173 | 0.04 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr5_-_140998481 | 0.04 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr1_+_198608146 | 0.04 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr20_+_5987890 | 0.04 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr10_-_21186144 | 0.03 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr1_+_193028717 | 0.03 |
ENST00000415442.2
ENST00000506303.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr1_-_47655686 | 0.03 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr1_-_44482979 | 0.03 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr8_-_18541603 | 0.03 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr12_+_109577202 | 0.03 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_+_40506255 | 0.03 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_-_79816965 | 0.03 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_-_109776901 | 0.03 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr8_-_131399110 | 0.03 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr17_-_33390667 | 0.03 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_-_58295712 | 0.03 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_-_115127611 | 0.02 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr6_-_111136299 | 0.02 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr9_-_14308004 | 0.02 |
ENST00000493697.1
|
NFIB
|
nuclear factor I/B |
| chr6_+_12717892 | 0.02 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr15_-_43877062 | 0.02 |
ENST00000381885.1
ENST00000396923.3 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr12_-_30887948 | 0.02 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr14_+_23727694 | 0.02 |
ENST00000399905.1
ENST00000470456.1 |
C14orf164
|
chromosome 14 open reading frame 164 |
| chr2_+_54951679 | 0.02 |
ENST00000356458.6
|
EML6
|
echinoderm microtubule associated protein like 6 |
| chrX_-_18690210 | 0.02 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr1_-_31666767 | 0.02 |
ENST00000530145.1
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr9_-_79520989 | 0.02 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chrX_+_139791917 | 0.02 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr6_-_139308777 | 0.02 |
ENST00000529597.1
ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr1_+_160370344 | 0.02 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr4_-_10686475 | 0.02 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr5_-_140998616 | 0.02 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr19_-_7697857 | 0.02 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr1_-_193028426 | 0.01 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr16_+_89228757 | 0.01 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr12_-_6233828 | 0.01 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr12_-_118797475 | 0.01 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_74421961 | 0.01 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr14_+_32030582 | 0.01 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chr22_-_28490123 | 0.01 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr5_+_66300446 | 0.01 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr4_-_47983519 | 0.01 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr3_+_159557637 | 0.01 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr5_+_66300464 | 0.01 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_39760129 | 0.01 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr6_+_4087664 | 0.01 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr17_-_9929581 | 0.01 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr2_-_158300556 | 0.01 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr13_-_36050819 | 0.01 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chr18_-_73967160 | 0.01 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr12_+_93130311 | 0.01 |
ENST00000344636.3
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr19_-_7698599 | 0.01 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr15_-_74421477 | 0.01 |
ENST00000514871.1
|
RP11-247C2.2
|
HCG2004779; Uncharacterized protein |
| chr11_+_31531291 | 0.01 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr1_-_190446759 | 0.01 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_-_109777128 | 0.01 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr10_+_123951957 | 0.01 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_49833978 | 0.01 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr8_-_49834299 | 0.01 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr6_+_130339710 | 0.00 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr7_+_142982023 | 0.00 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr9_+_125132803 | 0.00 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_115342349 | 0.00 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chrX_-_133931164 | 0.00 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr2_-_122262593 | 0.00 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr12_-_52779433 | 0.00 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr19_-_10446449 | 0.00 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr2_-_70780572 | 0.00 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr8_+_97773202 | 0.00 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr5_+_154393260 | 0.00 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr10_+_18549645 | 0.00 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_-_50419055 | 0.00 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_+_66820058 | 0.00 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_+_97773457 | 0.00 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr12_+_64798826 | 0.00 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr6_+_151042224 | 0.00 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_+_172309634 | 0.00 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr10_-_73848531 | 0.00 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.0 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.0 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |