Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
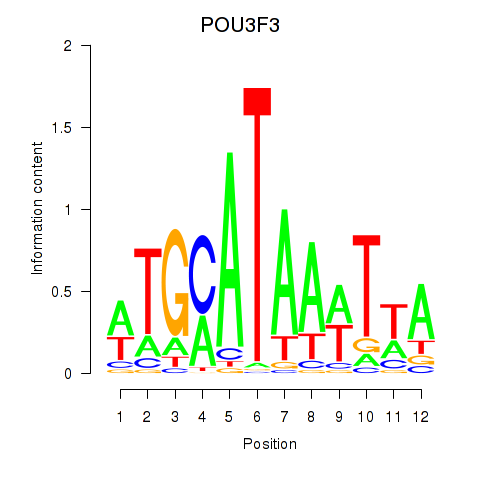
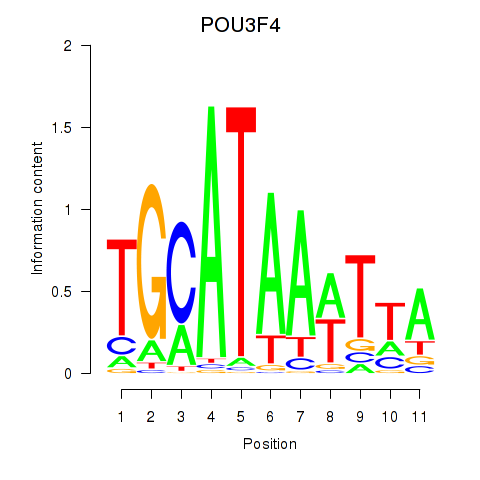
Results for POU3F3_POU3F4
Z-value: 1.39
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.99 | 7.2e-03 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_45670924 | 1.29 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr17_-_64216748 | 0.97 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr15_-_45670717 | 0.88 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr12_-_42631529 | 0.79 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr8_-_64080945 | 0.78 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr16_-_10652993 | 0.77 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr16_-_28192360 | 0.77 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr1_+_161035655 | 0.69 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr11_-_111649015 | 0.65 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr9_-_14322319 | 0.55 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr1_+_81106951 | 0.54 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr15_-_76352069 | 0.54 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr1_+_81001398 | 0.53 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_+_28099700 | 0.53 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr2_+_161993465 | 0.52 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr2_-_228244013 | 0.51 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr15_+_76352178 | 0.49 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr10_+_696000 | 0.48 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr11_-_118305921 | 0.48 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr6_-_134861089 | 0.48 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr1_+_209602156 | 0.47 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr2_+_40973618 | 0.46 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr17_-_39023462 | 0.43 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr7_-_50628745 | 0.42 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr1_-_151345159 | 0.41 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr10_-_90712520 | 0.40 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr13_-_46679144 | 0.39 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr3_+_151591422 | 0.35 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr9_-_15472730 | 0.35 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr4_-_110723335 | 0.34 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr12_-_111926342 | 0.34 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr19_-_18632861 | 0.34 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr13_+_108921977 | 0.34 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_+_38244638 | 0.33 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr16_-_30064244 | 0.32 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chrX_-_80457385 | 0.32 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr7_-_38948774 | 0.31 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr15_+_69857515 | 0.31 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr16_-_18887627 | 0.31 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr5_-_127418573 | 0.29 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr2_-_224702740 | 0.29 |
ENST00000444408.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr8_-_27695552 | 0.29 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr1_-_151882031 | 0.29 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr16_-_21314360 | 0.29 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr7_-_105332084 | 0.29 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr12_-_91573132 | 0.29 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chrX_+_78003204 | 0.28 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr3_-_148939598 | 0.28 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr7_-_229557 | 0.28 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr9_+_34458851 | 0.27 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr19_-_13900972 | 0.27 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr3_+_12525931 | 0.26 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr10_+_114710211 | 0.26 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_28099683 | 0.26 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr4_-_110723194 | 0.26 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr10_+_695888 | 0.25 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr2_+_120687335 | 0.25 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr4_-_122148620 | 0.24 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr2_-_152118276 | 0.24 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr10_+_114709999 | 0.24 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrM_+_5824 | 0.24 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr21_+_17443521 | 0.23 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr6_+_26273144 | 0.23 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr11_-_95657231 | 0.22 |
ENST00000409459.1
ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2
|
myotubularin related protein 2 |
| chr3_+_189507460 | 0.22 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr5_+_140174429 | 0.22 |
ENST00000520672.2
ENST00000378132.1 ENST00000526136.1 |
PCDHA2
|
protocadherin alpha 2 |
| chr16_-_745946 | 0.22 |
ENST00000562563.1
|
FBXL16
|
F-box and leucine-rich repeat protein 16 |
| chr9_+_108424738 | 0.21 |
ENST00000334077.3
|
TAL2
|
T-cell acute lymphocytic leukemia 2 |
| chr3_+_189507432 | 0.21 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr5_+_140753444 | 0.21 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr2_-_55237484 | 0.21 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr9_-_95244781 | 0.21 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr4_-_110723134 | 0.21 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr19_-_44123734 | 0.20 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr5_+_36152163 | 0.20 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_+_41221975 | 0.20 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chr3_-_196242233 | 0.20 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr13_-_26795840 | 0.19 |
ENST00000381570.3
ENST00000399762.2 ENST00000346166.3 |
RNF6
|
ring finger protein (C3H2C3 type) 6 |
| chr17_-_5321549 | 0.19 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr12_+_56623827 | 0.19 |
ENST00000424625.1
ENST00000419753.1 ENST00000454355.2 ENST00000417965.1 ENST00000436633.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr5_+_36152091 | 0.18 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr3_+_53528659 | 0.18 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr2_+_28974603 | 0.18 |
ENST00000441461.1
ENST00000358506.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_-_101034070 | 0.18 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr14_-_65409438 | 0.18 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr12_+_49297899 | 0.18 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr1_+_6105974 | 0.18 |
ENST00000378083.3
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr1_+_104198377 | 0.18 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr1_-_27701307 | 0.18 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr13_-_46679185 | 0.17 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr9_-_14180778 | 0.17 |
ENST00000380924.1
ENST00000543693.1 |
NFIB
|
nuclear factor I/B |
| chr2_+_33359646 | 0.17 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.17 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_190541153 | 0.17 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr17_-_38978847 | 0.16 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr8_-_124749609 | 0.16 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr3_+_189507523 | 0.16 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr1_+_73771844 | 0.16 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr21_+_17443434 | 0.16 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr19_-_44124019 | 0.16 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr9_+_21409146 | 0.16 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr2_-_239140011 | 0.16 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chr10_+_71561704 | 0.16 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr12_+_58003935 | 0.16 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr1_-_238108575 | 0.16 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr1_+_104159999 | 0.15 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr12_+_49740700 | 0.15 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chrX_-_39956656 | 0.15 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr4_+_102711764 | 0.15 |
ENST00000322953.4
ENST00000428908.1 |
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr7_+_150065278 | 0.15 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chrX_+_80457442 | 0.15 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr6_-_2634820 | 0.15 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr2_+_28974668 | 0.15 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr12_+_6554021 | 0.14 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr1_-_62190793 | 0.14 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr2_+_145780739 | 0.14 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr12_-_6960407 | 0.14 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr10_-_105110831 | 0.14 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr17_-_7387524 | 0.14 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr2_+_161993412 | 0.14 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr13_-_20110902 | 0.14 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr13_+_103459704 | 0.13 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr5_-_42812143 | 0.13 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_-_119634141 | 0.13 |
ENST00000409003.4
ENST00000526328.1 ENST00000314727.4 ENST00000526765.1 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr19_-_56135928 | 0.13 |
ENST00000591479.1
ENST00000325351.4 |
ZNF784
|
zinc finger protein 784 |
| chr2_-_58468437 | 0.13 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr2_-_46769694 | 0.13 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr1_-_243417762 | 0.13 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr2_-_204398141 | 0.12 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_-_32529973 | 0.12 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_-_183387430 | 0.12 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_+_73455788 | 0.12 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr8_+_52730143 | 0.12 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr3_-_141747459 | 0.12 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_82782952 | 0.11 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr5_+_140474181 | 0.11 |
ENST00000194155.4
|
PCDHB2
|
protocadherin beta 2 |
| chr4_-_152149033 | 0.11 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr11_-_28129656 | 0.11 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr12_+_56624436 | 0.11 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr3_-_112564797 | 0.11 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr7_+_143771275 | 0.11 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr6_-_136847099 | 0.11 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr7_-_140482926 | 0.11 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr6_+_74171301 | 0.11 |
ENST00000415954.2
ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1
|
mitochondrial tRNA translation optimization 1 |
| chr1_+_59775752 | 0.11 |
ENST00000371212.1
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr4_-_102268708 | 0.11 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_-_122107549 | 0.11 |
ENST00000355329.3
|
MORN3
|
MORN repeat containing 3 |
| chr14_-_65409502 | 0.11 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr1_+_61547894 | 0.11 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr1_+_113009163 | 0.10 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr10_-_48806939 | 0.10 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr14_+_24407940 | 0.10 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr12_+_50690489 | 0.10 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr1_+_78383813 | 0.10 |
ENST00000342754.5
|
NEXN
|
nexilin (F actin binding protein) |
| chr6_-_116833500 | 0.10 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr11_+_36616355 | 0.10 |
ENST00000532470.2
|
C11orf74
|
chromosome 11 open reading frame 74 |
| chr17_-_39156138 | 0.10 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr1_-_179112173 | 0.10 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_+_55600580 | 0.10 |
ENST00000457326.2
|
CAPNS2
|
calpain, small subunit 2 |
| chr22_-_39268308 | 0.10 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr22_-_43036607 | 0.10 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr17_-_38821373 | 0.10 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr12_+_10124001 | 0.09 |
ENST00000396507.3
ENST00000304361.4 ENST00000434319.2 |
CLEC12A
|
C-type lectin domain family 12, member A |
| chr3_-_167371740 | 0.09 |
ENST00000466760.1
ENST00000479765.1 |
WDR49
|
WD repeat domain 49 |
| chr10_-_116444371 | 0.09 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr12_-_120241187 | 0.09 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_1297157 | 0.09 |
ENST00000477278.2
|
MXRA8
|
matrix-remodelling associated 8 |
| chr4_-_168155730 | 0.09 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_-_39734783 | 0.09 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr15_+_52121822 | 0.09 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr7_+_77469439 | 0.09 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr6_-_136847610 | 0.09 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chrX_+_23682379 | 0.09 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr2_-_113993020 | 0.09 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr5_-_137374288 | 0.09 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr11_+_124824000 | 0.09 |
ENST00000529051.1
ENST00000344762.5 |
CCDC15
|
coiled-coil domain containing 15 |
| chr1_-_67142710 | 0.09 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr10_+_57358750 | 0.08 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr12_-_100486668 | 0.08 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr3_-_58613323 | 0.08 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr2_+_28974531 | 0.08 |
ENST00000420282.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_104239076 | 0.08 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr7_-_14880892 | 0.08 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr13_+_40229764 | 0.08 |
ENST00000416691.1
ENST00000422759.2 ENST00000455146.3 |
COG6
|
component of oligomeric golgi complex 6 |
| chr14_+_70918874 | 0.08 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr16_+_27279526 | 0.08 |
ENST00000566854.1
|
CTD-3203P2.2
|
HCG1815999; Uncharacterized protein |
| chr16_-_279405 | 0.08 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr1_-_179112189 | 0.07 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_-_26220366 | 0.07 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr1_+_112016414 | 0.07 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr6_-_49712147 | 0.07 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_+_49297887 | 0.07 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr11_+_3829691 | 0.07 |
ENST00000278243.4
ENST00000463452.2 ENST00000479072.1 ENST00000496834.2 ENST00000469307.2 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr4_-_144826682 | 0.07 |
ENST00000358615.4
ENST00000437468.2 |
GYPE
|
glycophorin E (MNS blood group) |
| chr22_+_45148432 | 0.07 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr1_-_2458026 | 0.07 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr5_-_36152031 | 0.07 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr3_+_35680339 | 0.07 |
ENST00000450234.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_130080977 | 0.07 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr12_+_64173888 | 0.07 |
ENST00000537373.1
|
TMEM5
|
transmembrane protein 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.1 | 0.6 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.4 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.5 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 0.2 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.3 | 1.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |