Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
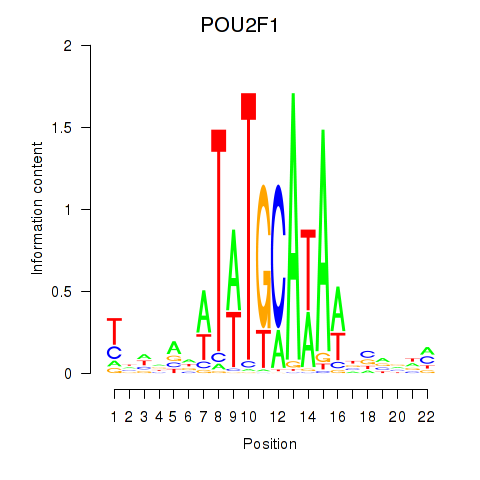
Results for POU2F1
Z-value: 0.67
Transcription factors associated with POU2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F1
|
ENSG00000143190.17 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F1 | hg19_v2_chr1_+_167190066_167190156 | 0.90 | 9.6e-02 | Click! |
Activity profile of POU2F1 motif
Sorted Z-values of POU2F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_100065440 | 0.49 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_+_107318157 | 0.44 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chrY_+_15418467 | 0.41 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr6_+_26124373 | 0.33 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr18_+_44812072 | 0.33 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr3_-_58200398 | 0.32 |
ENST00000318316.3
ENST00000460422.1 ENST00000483681.1 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr3_+_118892362 | 0.31 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr1_+_90308981 | 0.30 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr12_-_58329819 | 0.30 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr17_-_64225508 | 0.28 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr14_-_92247032 | 0.27 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr14_-_107283278 | 0.26 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr8_-_33370607 | 0.26 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr3_-_148939598 | 0.24 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr7_+_20686946 | 0.24 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr2_+_219524379 | 0.24 |
ENST00000443791.1
ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr12_-_110906027 | 0.24 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr3_-_165555200 | 0.23 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr2_+_183582774 | 0.23 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr3_-_148939835 | 0.23 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr4_-_100065419 | 0.22 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr2_-_152684977 | 0.21 |
ENST00000428992.2
ENST00000295087.8 |
ARL5A
|
ADP-ribosylation factor-like 5A |
| chr2_+_90458201 | 0.21 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr3_+_118892411 | 0.21 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr9_-_123812542 | 0.20 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr9_+_124088860 | 0.20 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr3_+_133292574 | 0.20 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chrX_+_47050798 | 0.20 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr8_+_27238147 | 0.20 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_196621002 | 0.20 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr6_-_26043885 | 0.20 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr9_-_19065082 | 0.19 |
ENST00000415524.1
|
HAUS6
|
HAUS augmin-like complex, subunit 6 |
| chr1_+_25598872 | 0.19 |
ENST00000328664.4
|
RHD
|
Rh blood group, D antigen |
| chr2_+_44001172 | 0.19 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr17_+_37844331 | 0.19 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr7_+_116451100 | 0.18 |
ENST00000464223.1
ENST00000484325.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr17_+_43239191 | 0.18 |
ENST00000589230.1
|
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr7_-_84121858 | 0.17 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr15_-_45670924 | 0.17 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_145940214 | 0.17 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr8_-_101730061 | 0.17 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr12_-_50616382 | 0.17 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr11_-_68611721 | 0.17 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr9_-_21335356 | 0.16 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr12_-_10978957 | 0.16 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr7_+_37723420 | 0.16 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr12_-_118796910 | 0.16 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr9_+_116638562 | 0.16 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr13_+_48611665 | 0.16 |
ENST00000258662.2
|
NUDT15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr4_-_152147579 | 0.15 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr15_-_55489097 | 0.15 |
ENST00000260443.4
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr1_+_100436065 | 0.15 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr1_-_101360374 | 0.15 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr1_-_101360331 | 0.15 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr17_-_37844267 | 0.15 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr14_+_19377522 | 0.15 |
ENST00000550708.1
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12 |
| chr1_+_62439037 | 0.14 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr8_-_117886563 | 0.14 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr13_+_108870714 | 0.14 |
ENST00000375898.3
|
ABHD13
|
abhydrolase domain containing 13 |
| chr17_-_41910505 | 0.14 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr6_-_15586238 | 0.14 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr11_-_61596753 | 0.14 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr5_+_94982558 | 0.13 |
ENST00000311364.4
ENST00000458310.1 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr1_-_17231271 | 0.13 |
ENST00000606899.1
|
RP11-108M9.6
|
RP11-108M9.6 |
| chr11_-_107729504 | 0.13 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr18_-_68004529 | 0.13 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr22_+_24030321 | 0.13 |
ENST00000401461.1
|
RGL4
|
ral guanine nucleotide dissociation stimulator-like 4 |
| chr19_+_17858547 | 0.13 |
ENST00000600676.1
ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1
|
FCH domain only 1 |
| chr5_+_94982435 | 0.13 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr8_-_117886612 | 0.12 |
ENST00000520992.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr19_+_14693888 | 0.12 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr2_-_113594279 | 0.12 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr11_-_58343319 | 0.12 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chrX_-_135962923 | 0.12 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr22_-_16449805 | 0.11 |
ENST00000252835.4
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1 |
| chr1_-_201123546 | 0.11 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr7_-_103848405 | 0.11 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chrX_+_119384607 | 0.11 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr5_+_140071178 | 0.11 |
ENST00000508522.1
ENST00000448069.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr1_-_201123586 | 0.11 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr4_-_110723134 | 0.11 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr10_-_29811456 | 0.11 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr15_-_63448973 | 0.10 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr15_-_50838888 | 0.10 |
ENST00000532404.1
|
USP50
|
ubiquitin specific peptidase 50 |
| chr19_+_17337027 | 0.10 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr3_+_151591422 | 0.10 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr18_+_61144160 | 0.10 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chrX_-_135962876 | 0.10 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chrX_-_130423240 | 0.10 |
ENST00000370910.1
ENST00000370901.4 |
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr3_-_145940126 | 0.10 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr1_-_101360205 | 0.10 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr9_-_35080013 | 0.09 |
ENST00000378643.3
|
FANCG
|
Fanconi anemia, complementation group G |
| chr1_-_106161540 | 0.09 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr5_+_118668846 | 0.09 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_+_122720681 | 0.09 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr3_-_155524049 | 0.09 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr12_-_118797475 | 0.09 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_27849378 | 0.09 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr22_+_18721427 | 0.09 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr11_+_94300474 | 0.09 |
ENST00000299001.6
|
PIWIL4
|
piwi-like RNA-mediated gene silencing 4 |
| chr5_+_140739537 | 0.09 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr14_-_21270995 | 0.09 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr15_-_89089860 | 0.09 |
ENST00000558413.1
ENST00000564406.1 ENST00000268148.8 |
DET1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr17_+_45728427 | 0.08 |
ENST00000540627.1
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr2_+_173292059 | 0.08 |
ENST00000412899.1
ENST00000409532.1 |
ITGA6
|
integrin, alpha 6 |
| chr15_+_48623208 | 0.08 |
ENST00000559935.1
ENST00000559416.1 |
DUT
|
deoxyuridine triphosphatase |
| chr12_-_118796971 | 0.08 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_6614565 | 0.08 |
ENST00000377705.5
|
NOL9
|
nucleolar protein 9 |
| chr3_+_149530836 | 0.08 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr15_-_45670717 | 0.08 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr12_-_11139511 | 0.08 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr19_-_9731872 | 0.08 |
ENST00000424629.1
ENST00000326044.5 ENST00000354661.4 ENST00000435550.1 ENST00000444611.1 ENST00000421525.1 |
ZNF561
|
zinc finger protein 561 |
| chr3_-_33686925 | 0.08 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_-_130423386 | 0.08 |
ENST00000370903.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr20_+_36888551 | 0.07 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr17_-_2304365 | 0.07 |
ENST00000575394.1
ENST00000174618.4 |
MNT
|
MAX network transcriptional repressor |
| chr1_-_241683001 | 0.07 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr9_-_39239171 | 0.07 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr4_-_110723194 | 0.07 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chrX_+_57313113 | 0.07 |
ENST00000374900.4
|
FAAH2
|
fatty acid amide hydrolase 2 |
| chr17_-_19364269 | 0.07 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr1_-_109935819 | 0.07 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr9_-_86593238 | 0.06 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr17_+_43239231 | 0.06 |
ENST00000591576.1
ENST00000591070.1 ENST00000592695.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr1_-_169703203 | 0.06 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr11_+_85359062 | 0.06 |
ENST00000532180.1
|
TMEM126A
|
transmembrane protein 126A |
| chr10_-_116286656 | 0.06 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr3_-_46249878 | 0.06 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr17_+_70026795 | 0.06 |
ENST00000472655.2
ENST00000538810.1 |
RP11-84E24.3
|
long intergenic non-protein coding RNA 1152 |
| chr2_-_44550441 | 0.06 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr17_+_7387677 | 0.06 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr4_-_110723335 | 0.06 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr11_-_107729287 | 0.06 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr11_-_14521379 | 0.05 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr12_+_40618873 | 0.05 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr14_-_68283291 | 0.05 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chrX_-_130423200 | 0.05 |
ENST00000361420.3
|
IGSF1
|
immunoglobulin superfamily, member 1 |
| chr19_+_17858509 | 0.05 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr3_+_32737027 | 0.05 |
ENST00000454516.2
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr10_+_13628921 | 0.05 |
ENST00000378572.3
|
PRPF18
|
pre-mRNA processing factor 18 |
| chr12_-_11062161 | 0.05 |
ENST00000390677.2
|
TAS2R13
|
taste receptor, type 2, member 13 |
| chr22_+_41697520 | 0.05 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr2_+_36923830 | 0.05 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr14_-_65346555 | 0.05 |
ENST00000542895.1
ENST00000556626.1 |
SPTB
|
spectrin, beta, erythrocytic |
| chr1_+_161736072 | 0.04 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr10_+_118187379 | 0.04 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr19_+_17337007 | 0.04 |
ENST00000215061.4
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr3_+_197464046 | 0.04 |
ENST00000428738.1
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr5_-_111093081 | 0.04 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr2_-_74007095 | 0.04 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr20_+_39657454 | 0.04 |
ENST00000361337.2
|
TOP1
|
topoisomerase (DNA) I |
| chr2_+_162087577 | 0.04 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_144940477 | 0.04 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr12_-_58329888 | 0.04 |
ENST00000546580.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr12_+_105380073 | 0.04 |
ENST00000552951.1
ENST00000280749.5 |
C12orf45
|
chromosome 12 open reading frame 45 |
| chr3_-_33686743 | 0.04 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr15_+_39542867 | 0.03 |
ENST00000318578.3
ENST00000561223.1 |
C15orf54
|
chromosome 15 open reading frame 54 |
| chr7_+_64838712 | 0.03 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr12_-_7596735 | 0.03 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr2_+_36923933 | 0.03 |
ENST00000497382.1
ENST00000404084.1 ENST00000379241.3 ENST00000401530.1 |
VIT
|
vitrin |
| chr6_-_11779403 | 0.03 |
ENST00000414691.3
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr8_+_86121448 | 0.03 |
ENST00000520225.1
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr15_-_55541227 | 0.03 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr3_-_121379739 | 0.03 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_+_111855414 | 0.03 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr5_+_67588391 | 0.03 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr21_-_33975547 | 0.03 |
ENST00000431599.1
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr18_-_69246186 | 0.03 |
ENST00000568095.1
|
RP11-510D19.1
|
RP11-510D19.1 |
| chr13_-_20110902 | 0.03 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr7_-_99149715 | 0.03 |
ENST00000449309.1
|
FAM200A
|
family with sequence similarity 200, member A |
| chr9_-_140196703 | 0.03 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr12_-_111358372 | 0.03 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr10_-_31288398 | 0.03 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr16_+_81070792 | 0.03 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr11_-_58980342 | 0.03 |
ENST00000361050.3
|
MPEG1
|
macrophage expressed 1 |
| chr17_-_41623716 | 0.03 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr16_-_90142338 | 0.03 |
ENST00000407825.1
ENST00000449207.2 |
PRDM7
|
PR domain containing 7 |
| chr14_-_21270561 | 0.03 |
ENST00000412779.2
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr5_-_1551802 | 0.03 |
ENST00000503113.1
|
CTD-2245E15.3
|
CTD-2245E15.3 |
| chr2_-_145275109 | 0.02 |
ENST00000431672.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_66951382 | 0.02 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr20_+_5731083 | 0.02 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr6_-_11779014 | 0.02 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr4_+_158493642 | 0.02 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr1_+_146373546 | 0.02 |
ENST00000446760.2
|
NBPF12
|
neuroblastoma breakpoint family, member 12 |
| chr9_+_134065519 | 0.02 |
ENST00000531600.1
|
NUP214
|
nucleoporin 214kDa |
| chr10_-_115904361 | 0.02 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr22_-_29949657 | 0.02 |
ENST00000428374.1
|
THOC5
|
THO complex 5 |
| chr2_-_145275211 | 0.02 |
ENST00000462355.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_204571198 | 0.02 |
ENST00000374481.3
ENST00000458610.2 ENST00000324106.8 |
CD28
|
CD28 molecule |
| chr8_-_117768023 | 0.02 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr3_+_149531607 | 0.02 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr6_+_55192267 | 0.02 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr15_-_22473353 | 0.02 |
ENST00000557788.2
|
IGHV4OR15-8
|
immunoglobulin heavy variable 4/OR15-8 (non-functional) |
| chr4_-_153274078 | 0.02 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_+_106631966 | 0.02 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr17_-_29624343 | 0.02 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr6_-_152489484 | 0.01 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr4_+_146539415 | 0.01 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr6_-_35992270 | 0.01 |
ENST00000394602.2
ENST00000355574.2 |
SLC26A8
|
solute carrier family 26 (anion exchanger), member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.2 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 0.2 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.4 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.0 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.1 | 0.4 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.2 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.2 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |