Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
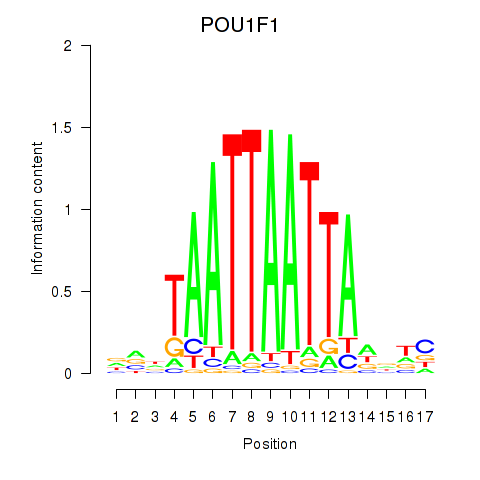
Results for POU1F1
Z-value: 0.30
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325612_87325654 | 0.97 | 2.6e-02 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_76251912 | 0.48 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr17_-_64225508 | 0.41 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr6_+_26087509 | 0.32 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr8_-_124741451 | 0.30 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr2_+_161993465 | 0.26 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_+_26104104 | 0.23 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr1_-_197036364 | 0.23 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chrX_-_71458802 | 0.23 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr8_+_98900132 | 0.22 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_-_35013217 | 0.21 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr11_+_327171 | 0.21 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr7_-_140482926 | 0.21 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr20_+_31870927 | 0.21 |
ENST00000253354.1
|
BPIFB1
|
BPI fold containing family B, member 1 |
| chr16_+_12059091 | 0.21 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr7_-_111032971 | 0.19 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr18_-_59274139 | 0.19 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chrX_+_43515467 | 0.18 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr6_+_26087646 | 0.18 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr5_+_111755280 | 0.17 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr19_+_49199209 | 0.17 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr17_+_40950797 | 0.17 |
ENST00000588408.1
ENST00000585355.1 |
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr5_-_111754948 | 0.17 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr4_+_95128748 | 0.17 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_113568207 | 0.17 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr2_-_152118276 | 0.16 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr12_-_25150409 | 0.16 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr19_+_52873166 | 0.16 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr6_-_52859046 | 0.16 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_62538089 | 0.16 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr21_-_39705323 | 0.15 |
ENST00000436845.1
|
AP001422.3
|
AP001422.3 |
| chr3_+_160939050 | 0.15 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr22_+_23487513 | 0.15 |
ENST00000263116.2
ENST00000341989.4 |
RAB36
|
RAB36, member RAS oncogene family |
| chr2_-_14541060 | 0.15 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr3_+_138340067 | 0.14 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr6_-_109702885 | 0.14 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr12_+_69753448 | 0.14 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr19_-_36304201 | 0.14 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr15_-_56757329 | 0.13 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr7_-_16872932 | 0.13 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr3_+_151591422 | 0.13 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chrX_+_154113317 | 0.12 |
ENST00000354461.2
|
H2AFB1
|
H2A histone family, member B1 |
| chr10_+_94352956 | 0.12 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr15_+_58702742 | 0.12 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr14_+_55494323 | 0.12 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr14_+_55493920 | 0.12 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr17_+_1674982 | 0.12 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_-_4938712 | 0.12 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr16_+_21623392 | 0.12 |
ENST00000562961.1
|
METTL9
|
methyltransferase like 9 |
| chr2_+_161993412 | 0.12 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_154689596 | 0.11 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr12_-_42631529 | 0.10 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr11_-_111649015 | 0.10 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_17981462 | 0.10 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr14_-_92247032 | 0.10 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr5_+_115177178 | 0.10 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr2_+_232575168 | 0.10 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr7_+_12544025 | 0.10 |
ENST00000443874.1
ENST00000424453.1 |
AC005281.1
|
AC005281.1 |
| chr5_-_125930929 | 0.10 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr7_+_13141010 | 0.10 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr5_+_95066823 | 0.10 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr21_+_35553045 | 0.10 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr10_+_695888 | 0.10 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr4_+_41937131 | 0.09 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chrX_-_15288154 | 0.09 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr3_+_28390637 | 0.09 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr10_+_696000 | 0.09 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr7_+_115862858 | 0.09 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chrX_+_154610428 | 0.09 |
ENST00000354514.4
|
H2AFB2
|
H2A histone family, member B2 |
| chrX_+_56590002 | 0.09 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr19_-_51920952 | 0.09 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr14_+_20187174 | 0.09 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr2_+_44001172 | 0.09 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr20_-_7921090 | 0.08 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_+_221051699 | 0.08 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr2_+_67624430 | 0.08 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr11_-_4719072 | 0.08 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr10_-_27529486 | 0.08 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr15_-_75748143 | 0.08 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_+_11333245 | 0.08 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr17_+_29664830 | 0.08 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr3_+_108308845 | 0.08 |
ENST00000479138.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr11_-_117186946 | 0.08 |
ENST00000313005.6
ENST00000528053.1 |
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr2_-_190044480 | 0.08 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr10_+_116697946 | 0.08 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr6_-_26235206 | 0.08 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr14_-_90420862 | 0.08 |
ENST00000556005.1
ENST00000555872.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr22_+_31460091 | 0.08 |
ENST00000432777.1
ENST00000422839.1 |
SMTN
|
smoothelin |
| chr10_+_17270214 | 0.07 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr1_+_81106951 | 0.07 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_+_43613566 | 0.07 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr3_-_194072019 | 0.07 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr2_+_242913327 | 0.07 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chrX_-_102943022 | 0.07 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chrX_+_150565653 | 0.07 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr19_-_4535233 | 0.07 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr11_+_115498761 | 0.07 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr4_+_37455536 | 0.07 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr4_-_103746683 | 0.06 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_145940214 | 0.06 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr6_+_29555683 | 0.06 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr19_+_50016411 | 0.06 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_-_25874440 | 0.06 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr10_+_94594351 | 0.06 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr3_-_96337000 | 0.06 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr1_+_81771806 | 0.06 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr5_+_156696362 | 0.06 |
ENST00000377576.3
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr6_-_134861089 | 0.06 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr6_+_28092338 | 0.06 |
ENST00000340487.4
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr14_+_61449076 | 0.06 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr3_+_136649311 | 0.06 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr6_-_52859968 | 0.06 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr3_-_27498235 | 0.06 |
ENST00000295736.5
ENST00000428386.1 ENST00000428179.1 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_-_109584608 | 0.06 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr1_+_111682058 | 0.06 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr16_-_66764119 | 0.06 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr16_-_2379688 | 0.06 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr8_-_143859197 | 0.06 |
ENST00000395192.2
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr3_+_193310918 | 0.06 |
ENST00000361908.3
ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1
|
optic atrophy 1 (autosomal dominant) |
| chr4_-_185275104 | 0.06 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr12_-_10978957 | 0.06 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr11_-_22647350 | 0.06 |
ENST00000327470.3
|
FANCF
|
Fanconi anemia, complementation group F |
| chr20_+_5987890 | 0.05 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr1_+_43613612 | 0.05 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr7_+_130126165 | 0.05 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr10_+_51572408 | 0.05 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr4_+_48833234 | 0.05 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr4_+_119809984 | 0.05 |
ENST00000307142.4
ENST00000448416.2 ENST00000429713.2 |
SYNPO2
|
synaptopodin 2 |
| chr2_-_43266680 | 0.05 |
ENST00000425212.1
ENST00000422351.1 ENST00000449766.1 |
AC016735.2
|
AC016735.2 |
| chr9_+_125133315 | 0.05 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_76452097 | 0.05 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr16_-_28634874 | 0.05 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_-_14992264 | 0.05 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr6_+_26440700 | 0.05 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr20_+_5986727 | 0.05 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chrX_-_102942961 | 0.05 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr17_+_45728427 | 0.05 |
ENST00000540627.1
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr1_-_54411255 | 0.05 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr14_+_62164340 | 0.05 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr19_+_19144384 | 0.05 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr20_-_43589109 | 0.05 |
ENST00000372813.3
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr9_-_95055956 | 0.05 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr10_-_14596140 | 0.05 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr17_+_25799008 | 0.05 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr1_+_76251879 | 0.05 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr17_-_39165366 | 0.05 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr6_+_130339710 | 0.04 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr12_-_7596735 | 0.04 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr17_-_5321549 | 0.04 |
ENST00000572809.1
|
NUP88
|
nucleoporin 88kDa |
| chr6_+_26365443 | 0.04 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr10_-_115904361 | 0.04 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr8_-_57233103 | 0.04 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr17_+_78518617 | 0.04 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr10_-_13350447 | 0.04 |
ENST00000429930.1
|
AL138764.1
|
Uncharacterized protein |
| chr4_+_96012614 | 0.04 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr16_-_28937027 | 0.04 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr11_+_110300607 | 0.04 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr12_-_11287243 | 0.04 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr10_+_122610687 | 0.04 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr9_-_95056010 | 0.04 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr7_+_130126012 | 0.04 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr17_-_38821373 | 0.04 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr6_+_64345698 | 0.04 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr9_-_5833027 | 0.04 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr4_+_186990298 | 0.04 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr1_-_53608249 | 0.04 |
ENST00000371494.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr1_+_93297622 | 0.04 |
ENST00000315741.5
|
RPL5
|
ribosomal protein L5 |
| chr4_-_118006697 | 0.04 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr4_-_85654615 | 0.04 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr16_+_15489629 | 0.04 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr11_+_100784231 | 0.03 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr1_+_66820058 | 0.03 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr8_-_141774467 | 0.03 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr5_+_59783941 | 0.03 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr4_+_119810134 | 0.03 |
ENST00000434046.2
|
SYNPO2
|
synaptopodin 2 |
| chr16_-_18923035 | 0.03 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr9_-_85882145 | 0.03 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr5_+_60933634 | 0.03 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr19_+_50016610 | 0.03 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr8_+_67687413 | 0.03 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr13_-_49107303 | 0.03 |
ENST00000344532.3
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr3_+_195447738 | 0.03 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr11_+_59705928 | 0.03 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr10_+_57358750 | 0.03 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr6_-_161695074 | 0.03 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr16_+_48278178 | 0.03 |
ENST00000285737.4
ENST00000535754.1 |
LONP2
|
lon peptidase 2, peroxisomal |
| chr3_+_169539710 | 0.03 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chrX_+_84258832 | 0.03 |
ENST00000373173.2
|
APOOL
|
apolipoprotein O-like |
| chr2_+_172309634 | 0.03 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr12_+_64798095 | 0.03 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_+_65843130 | 0.03 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr15_-_65321943 | 0.03 |
ENST00000220058.4
|
MTFMT
|
mitochondrial methionyl-tRNA formyltransferase |
| chr11_+_4664650 | 0.03 |
ENST00000396952.5
|
OR51E1
|
olfactory receptor, family 51, subfamily E, member 1 |
| chr12_-_86650077 | 0.03 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr6_-_136788001 | 0.03 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr16_-_67217844 | 0.03 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr2_-_152118352 | 0.03 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr6_+_4087664 | 0.03 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr9_+_44868935 | 0.03 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr10_-_75226166 | 0.03 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr2_-_231989808 | 0.02 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.0 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.5 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.1 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |