Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
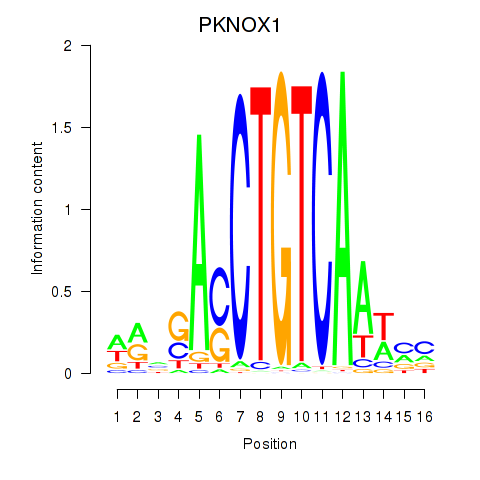
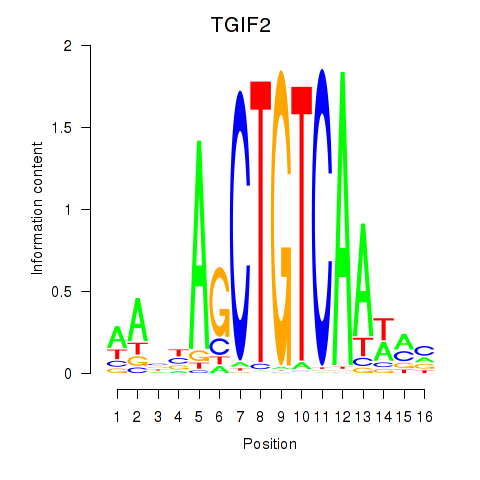
Results for PKNOX1_TGIF2
Z-value: 0.94
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | 0.91 | 9.1e-02 | Click! |
| TGIF2 | hg19_v2_chr20_+_35202909_35203075 | -0.57 | 4.3e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_63989004 | 1.05 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr12_+_62654155 | 0.84 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr1_-_63988846 | 0.77 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr18_-_52626622 | 0.67 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr12_+_95611536 | 0.65 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr3_-_52719888 | 0.64 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr9_+_111696664 | 0.64 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr7_+_76139741 | 0.63 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr11_+_6411636 | 0.59 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr2_-_203103185 | 0.52 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr4_-_175443788 | 0.51 |
ENST00000541923.1
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr6_+_26124373 | 0.51 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr18_-_54318353 | 0.50 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr11_+_827553 | 0.48 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr2_+_191208601 | 0.47 |
ENST00000413239.1
ENST00000431594.1 ENST00000444194.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr1_-_247171347 | 0.46 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr17_-_64225508 | 0.46 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr10_-_118764862 | 0.41 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr7_+_79765071 | 0.38 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr7_+_76139833 | 0.37 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr9_-_123605177 | 0.37 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr11_-_67415048 | 0.36 |
ENST00000529256.1
|
ACY3
|
aspartoacylase (aminocyclase) 3 |
| chr3_-_3221358 | 0.36 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr15_-_37391614 | 0.36 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr3_-_142166904 | 0.35 |
ENST00000264951.4
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr17_+_68071389 | 0.35 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_-_8318847 | 0.35 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr6_-_160209471 | 0.34 |
ENST00000539948.1
|
TCP1
|
t-complex 1 |
| chr1_+_246729815 | 0.34 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr6_+_111195973 | 0.33 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr8_-_99837856 | 0.32 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr4_+_128886584 | 0.32 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr8_+_72755367 | 0.32 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr9_-_132805430 | 0.32 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr15_-_76352069 | 0.32 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr9_-_33447584 | 0.32 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr5_+_173763250 | 0.31 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr16_+_2588012 | 0.31 |
ENST00000354836.5
ENST00000389224.3 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr1_+_24286287 | 0.30 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr4_+_166300084 | 0.29 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr5_-_55008072 | 0.29 |
ENST00000512208.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr21_+_35553045 | 0.28 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr1_+_104104379 | 0.28 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr1_-_54872059 | 0.28 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr2_-_61765732 | 0.28 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr12_+_69201923 | 0.28 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr17_+_78518617 | 0.27 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr3_-_52719912 | 0.27 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr7_-_156685841 | 0.27 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr11_+_6411670 | 0.27 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr2_+_118846008 | 0.27 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr15_+_96904487 | 0.26 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr15_-_37392086 | 0.25 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr3_-_142166796 | 0.25 |
ENST00000392981.2
|
XRN1
|
5'-3' exoribonuclease 1 |
| chr15_+_49913175 | 0.25 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr8_+_97657449 | 0.25 |
ENST00000220763.5
|
CPQ
|
carboxypeptidase Q |
| chr15_+_43477455 | 0.25 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr11_+_60552797 | 0.24 |
ENST00000308287.1
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10 |
| chr5_+_43603229 | 0.24 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr2_-_61765315 | 0.24 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr7_-_104909435 | 0.23 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr16_+_2587998 | 0.23 |
ENST00000441549.3
ENST00000268673.7 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr14_-_94856987 | 0.23 |
ENST00000449399.3
ENST00000404814.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr17_-_70053866 | 0.22 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr11_+_120207787 | 0.22 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr18_-_54305658 | 0.22 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr14_+_103566481 | 0.22 |
ENST00000380069.3
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr7_-_156803329 | 0.21 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr4_-_114682224 | 0.21 |
ENST00000342666.5
ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr3_-_33481835 | 0.21 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr9_-_123639445 | 0.21 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr15_+_49913201 | 0.21 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr10_-_11574274 | 0.21 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr6_-_131949200 | 0.21 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr1_+_154966058 | 0.21 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr14_-_94857004 | 0.21 |
ENST00000557492.1
ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_12512062 | 0.21 |
ENST00000595766.1
ENST00000430385.3 |
ZNF799
|
zinc finger protein 799 |
| chr12_+_10658489 | 0.21 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chrX_+_16804544 | 0.20 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr1_+_186649754 | 0.20 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr5_-_154230130 | 0.20 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr10_+_28822636 | 0.20 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr8_+_97506033 | 0.20 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr3_-_52488048 | 0.20 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr10_+_49514698 | 0.20 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr2_+_191208196 | 0.19 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr10_-_124768300 | 0.19 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr3_-_169587621 | 0.19 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr8_-_17752912 | 0.19 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chr2_+_233925064 | 0.19 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr4_+_57774042 | 0.18 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr2_-_74780176 | 0.18 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr9_-_100459639 | 0.18 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr6_-_131949305 | 0.17 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr5_-_81046904 | 0.17 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr12_-_31882027 | 0.17 |
ENST00000541931.1
ENST00000535408.1 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr10_-_118765081 | 0.17 |
ENST00000392903.2
ENST00000355371.4 |
KIAA1598
|
KIAA1598 |
| chr12_-_31881944 | 0.17 |
ENST00000537562.1
ENST00000537960.1 ENST00000536761.1 ENST00000542781.1 ENST00000457428.2 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr9_-_6470375 | 0.17 |
ENST00000355513.4
|
C9orf38
|
chromosome 9 open reading frame 38 |
| chr17_+_68071458 | 0.17 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr21_-_34144157 | 0.17 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr15_+_36871983 | 0.17 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_+_203765437 | 0.17 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr5_-_74348371 | 0.17 |
ENST00000503568.1
|
RP11-229C3.2
|
RP11-229C3.2 |
| chr12_+_11081828 | 0.17 |
ENST00000381847.3
ENST00000396400.3 |
PRH2
|
proline-rich protein HaeIII subfamily 2 |
| chr2_+_217082311 | 0.17 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr17_-_29151686 | 0.17 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr12_-_104531785 | 0.17 |
ENST00000551727.1
|
NFYB
|
nuclear transcription factor Y, beta |
| chr8_-_17555164 | 0.16 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr2_-_203103281 | 0.16 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr12_+_69186125 | 0.16 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr19_-_45663408 | 0.16 |
ENST00000317951.4
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr19_-_46145696 | 0.16 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr13_+_73302047 | 0.15 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr17_+_39346139 | 0.15 |
ENST00000398470.1
ENST00000318329.5 |
KRTAP9-1
|
keratin associated protein 9-1 |
| chr4_+_8183793 | 0.15 |
ENST00000509119.1
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr15_+_68346501 | 0.15 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr21_+_35552978 | 0.15 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr8_-_141774467 | 0.15 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr4_-_83483094 | 0.15 |
ENST00000508701.1
ENST00000454948.3 |
TMEM150C
|
transmembrane protein 150C |
| chr4_-_83483360 | 0.15 |
ENST00000449862.2
|
TMEM150C
|
transmembrane protein 150C |
| chr15_+_43477580 | 0.14 |
ENST00000356633.5
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr10_+_114710516 | 0.14 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_121734489 | 0.14 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr16_+_2587965 | 0.14 |
ENST00000342085.4
ENST00000566659.1 |
PDPK1
|
3-phosphoinositide dependent protein kinase-1 |
| chr19_-_12551849 | 0.14 |
ENST00000595562.1
ENST00000301547.5 |
CTD-3105H18.16
ZNF443
|
Uncharacterized protein zinc finger protein 443 |
| chr8_+_72755796 | 0.14 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr3_+_38017264 | 0.14 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_-_30457048 | 0.14 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr5_-_81046922 | 0.13 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_+_54173227 | 0.13 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr6_-_101329191 | 0.13 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr14_-_94759408 | 0.13 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr19_+_11877838 | 0.13 |
ENST00000357901.4
ENST00000454339.2 |
ZNF441
|
zinc finger protein 441 |
| chr4_+_48485341 | 0.13 |
ENST00000273861.4
|
SLC10A4
|
solute carrier family 10, member 4 |
| chr4_-_140477928 | 0.13 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_+_94536898 | 0.13 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_-_3504587 | 0.13 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr7_+_39605966 | 0.13 |
ENST00000223273.2
ENST00000448268.1 ENST00000432096.2 |
YAE1D1
|
Yae1 domain containing 1 |
| chr6_-_41673552 | 0.13 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr3_-_149093499 | 0.12 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr5_-_52405564 | 0.12 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr14_-_94759361 | 0.12 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr6_-_135818844 | 0.12 |
ENST00000524469.1
ENST00000367800.4 ENST00000327035.6 ENST00000457866.2 ENST00000265602.6 |
AHI1
|
Abelson helper integration site 1 |
| chr10_+_51565188 | 0.12 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr12_-_31479045 | 0.12 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr1_-_242612779 | 0.12 |
ENST00000427495.1
|
PLD5
|
phospholipase D family, member 5 |
| chr9_-_99180597 | 0.12 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr4_-_175443943 | 0.12 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr16_-_20817753 | 0.12 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr6_-_35656685 | 0.12 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr11_-_8832521 | 0.12 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr5_+_118406796 | 0.11 |
ENST00000503802.1
|
DMXL1
|
Dmx-like 1 |
| chr21_-_34143971 | 0.11 |
ENST00000290178.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr13_+_76413852 | 0.11 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr3_+_45636219 | 0.11 |
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1 |
| chr9_-_123638633 | 0.11 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr21_-_30257669 | 0.11 |
ENST00000303775.5
ENST00000351429.3 |
N6AMT1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr9_-_123639304 | 0.11 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr7_+_38217920 | 0.11 |
ENST00000396013.1
ENST00000440144.1 ENST00000453225.1 ENST00000429075.1 |
STARD3NL
|
STARD3 N-terminal like |
| chr12_+_95611569 | 0.11 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_+_8316442 | 0.11 |
ENST00000582812.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr13_-_41837620 | 0.11 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr6_+_112375462 | 0.11 |
ENST00000361714.1
|
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr14_+_61995722 | 0.11 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr7_+_73442487 | 0.11 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr7_+_106810165 | 0.11 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr15_-_31283798 | 0.11 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr9_-_139345270 | 0.11 |
ENST00000398335.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_-_225615599 | 0.11 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr9_-_86432547 | 0.10 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr2_-_18770812 | 0.10 |
ENST00000359846.2
ENST00000304081.4 ENST00000600945.1 ENST00000532967.1 ENST00000444297.2 |
NT5C1B
NT5C1B-RDH14
|
5'-nucleotidase, cytosolic IB NT5C1B-RDH14 readthrough |
| chr12_+_69004619 | 0.10 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr19_+_11750566 | 0.10 |
ENST00000344893.3
|
ZNF833P
|
zinc finger protein 833, pseudogene |
| chr3_+_63897605 | 0.10 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr7_+_38217818 | 0.10 |
ENST00000009041.7
ENST00000544203.1 ENST00000434197.1 |
STARD3NL
|
STARD3 N-terminal like |
| chr14_+_101359265 | 0.10 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr17_-_77967433 | 0.10 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr5_-_132073111 | 0.10 |
ENST00000403231.1
|
KIF3A
|
kinesin family member 3A |
| chr6_-_32920794 | 0.10 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr17_-_42767115 | 0.10 |
ENST00000315286.8
ENST00000588210.1 ENST00000457422.2 |
CCDC43
|
coiled-coil domain containing 43 |
| chr2_-_190446738 | 0.10 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_-_152589670 | 0.10 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr16_+_16484691 | 0.10 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr14_-_94856951 | 0.10 |
ENST00000553327.1
ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr10_+_94608245 | 0.10 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr4_+_123653807 | 0.10 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_-_70671216 | 0.10 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr9_-_115653176 | 0.10 |
ENST00000374228.4
|
SLC46A2
|
solute carrier family 46, member 2 |
| chr15_-_77197620 | 0.09 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chrX_-_148713365 | 0.09 |
ENST00000511776.1
ENST00000507237.1 |
TMEM185A
|
transmembrane protein 185A |
| chr4_+_88928777 | 0.09 |
ENST00000237596.2
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant) |
| chr4_-_140477910 | 0.09 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_+_28844648 | 0.09 |
ENST00000373832.1
ENST00000373831.3 |
RCC1
|
regulator of chromosome condensation 1 |
| chr2_-_86790472 | 0.09 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr1_-_186649543 | 0.09 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_+_23637763 | 0.09 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr14_+_62164340 | 0.09 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr2_+_198380289 | 0.09 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr17_+_75396637 | 0.09 |
ENST00000590825.1
|
SEPT9
|
septin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.7 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.1 | 0.6 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.3 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 | 0.3 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.7 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.2 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.5 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:1904048 | positive regulation of long term synaptic depression(GO:1900454) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.4 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.1 | GO:0072194 | detection of endogenous stimulus(GO:0009726) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.3 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.0 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.0 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 0.7 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.2 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.1 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.3 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.7 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |