Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
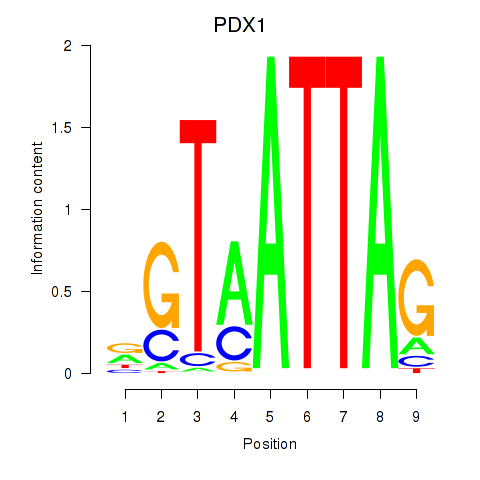
Results for PDX1
Z-value: 1.33
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | -0.69 | 3.1e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92777606 | 1.60 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr11_+_4510109 | 1.30 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chr8_-_42234745 | 1.27 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr7_+_37723336 | 1.22 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr1_+_158979792 | 1.09 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr15_+_63188009 | 1.08 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr2_+_66918558 | 0.97 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr7_+_129015484 | 0.89 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr14_+_61654271 | 0.87 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr14_-_77889860 | 0.86 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr2_+_228736335 | 0.86 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr6_-_31107127 | 0.83 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr4_-_41884620 | 0.83 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr17_-_71223839 | 0.79 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr15_+_91416092 | 0.74 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_-_108248169 | 0.74 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr4_+_169013666 | 0.73 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr20_+_45947246 | 0.69 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr11_+_35222629 | 0.68 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr3_-_121740969 | 0.67 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_106146547 | 0.66 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr11_-_8290263 | 0.66 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr11_+_110001723 | 0.63 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr6_+_126221034 | 0.62 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr17_-_46690839 | 0.61 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr17_+_72427477 | 0.61 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_190927447 | 0.60 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr12_-_54813229 | 0.60 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr5_-_141249154 | 0.57 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr15_+_80351910 | 0.56 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr1_-_190446759 | 0.55 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr11_-_8285405 | 0.54 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr13_-_46716969 | 0.54 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr18_-_47792851 | 0.53 |
ENST00000398545.4
|
CCDC11
|
coiled-coil domain containing 11 |
| chr20_+_60174827 | 0.52 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr10_-_21186144 | 0.52 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr11_-_115375107 | 0.51 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr7_+_129015671 | 0.48 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chrX_+_591524 | 0.44 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr2_+_234959376 | 0.44 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr17_-_8301132 | 0.44 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr2_+_87769459 | 0.44 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr15_+_62853562 | 0.43 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr5_+_135394840 | 0.43 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr1_+_107683644 | 0.43 |
ENST00000370067.1
|
NTNG1
|
netrin G1 |
| chrX_+_10126488 | 0.42 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr19_-_7968427 | 0.42 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr8_+_77593448 | 0.42 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr2_+_45168875 | 0.40 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr1_+_66796401 | 0.40 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr10_-_23633720 | 0.39 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr21_+_30673091 | 0.39 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr18_+_3447572 | 0.39 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_145277569 | 0.39 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr12_-_109221160 | 0.39 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr15_-_55562479 | 0.38 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_-_18430160 | 0.38 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr19_-_7697857 | 0.38 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr12_+_42725554 | 0.37 |
ENST00000546750.1
ENST00000547847.1 |
PPHLN1
|
periphilin 1 |
| chrX_-_106243294 | 0.37 |
ENST00000255495.7
|
MORC4
|
MORC family CW-type zinc finger 4 |
| chr12_+_64798826 | 0.37 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr11_+_65554493 | 0.37 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr4_-_70518941 | 0.36 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr4_+_30721968 | 0.35 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chrX_-_110655306 | 0.35 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr7_-_33102338 | 0.34 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr10_+_24738355 | 0.34 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr15_-_100882191 | 0.34 |
ENST00000268070.4
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr14_-_36988882 | 0.34 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr20_-_56286479 | 0.33 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_+_198608146 | 0.33 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr7_-_33102399 | 0.33 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr1_+_19967014 | 0.33 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr14_-_61116168 | 0.31 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr6_+_34204642 | 0.31 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr5_+_174151536 | 0.30 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr17_-_38928414 | 0.30 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr7_-_99716952 | 0.30 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr9_-_117267717 | 0.29 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr7_-_99717463 | 0.29 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_+_9953214 | 0.29 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr19_+_11071546 | 0.29 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr1_-_94147385 | 0.28 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_-_56321649 | 0.27 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr20_-_50418947 | 0.27 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr9_-_16728161 | 0.27 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr7_-_100860851 | 0.26 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr3_-_101039402 | 0.26 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr1_-_149859466 | 0.26 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr3_-_141747950 | 0.25 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_85430356 | 0.25 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_169396646 | 0.25 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr3_-_100565249 | 0.25 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_199996702 | 0.25 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr12_-_16761007 | 0.25 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr7_+_114055052 | 0.24 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr4_-_119759795 | 0.24 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr6_+_39760129 | 0.24 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr11_-_26593649 | 0.24 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr1_-_1709845 | 0.24 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr12_-_32908809 | 0.23 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr1_+_62439037 | 0.23 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_212965104 | 0.23 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr7_-_14028488 | 0.22 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr8_-_42358742 | 0.22 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr12_-_53171128 | 0.22 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr4_-_140544386 | 0.22 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr13_+_102104952 | 0.22 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_-_103749179 | 0.22 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_106243451 | 0.21 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr21_+_30672433 | 0.21 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chrX_-_19817869 | 0.21 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr15_+_41913690 | 0.21 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr3_+_69985792 | 0.21 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_+_72052983 | 0.20 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr18_+_55888767 | 0.20 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_+_107683436 | 0.20 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
| chr4_+_95916947 | 0.20 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr3_+_115342349 | 0.20 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr1_+_1846519 | 0.20 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr1_+_36621529 | 0.19 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_-_16762971 | 0.19 |
ENST00000540590.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr9_+_131062367 | 0.19 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr7_+_100860949 | 0.19 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr8_+_9953061 | 0.19 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr13_+_78315528 | 0.18 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chrX_-_13835147 | 0.18 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr12_-_21928515 | 0.18 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chrX_-_46187069 | 0.18 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_-_6233828 | 0.18 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr2_-_99871570 | 0.18 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr5_-_54988559 | 0.18 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr19_-_12889226 | 0.18 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr4_+_144354644 | 0.17 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr8_+_32579341 | 0.17 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr14_-_95236551 | 0.17 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr15_+_28624878 | 0.17 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr1_+_160370344 | 0.17 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr1_-_201140673 | 0.17 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr5_-_140998616 | 0.17 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr18_+_32173276 | 0.17 |
ENST00000591816.1
ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA
|
dystrobrevin, alpha |
| chr3_-_38071122 | 0.17 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr21_+_29911640 | 0.17 |
ENST00000412526.1
ENST00000455939.1 |
LINC00161
|
long intergenic non-protein coding RNA 161 |
| chr14_-_78083112 | 0.16 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr13_+_102104980 | 0.16 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_-_25042231 | 0.16 |
ENST00000510784.2
|
FAM65B
|
family with sequence similarity 65, member B |
| chr13_-_99667960 | 0.16 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr17_-_9929581 | 0.16 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr7_-_81399438 | 0.16 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr20_-_50419055 | 0.16 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr15_+_48498480 | 0.16 |
ENST00000380993.3
ENST00000396577.3 |
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr5_-_96478466 | 0.16 |
ENST00000274382.4
|
LIX1
|
Lix1 homolog (chicken) |
| chr18_+_59000815 | 0.16 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr1_-_232651312 | 0.16 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chrX_-_18690210 | 0.15 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr2_-_145188137 | 0.15 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr15_-_55563072 | 0.15 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr20_-_49307897 | 0.15 |
ENST00000535356.1
|
FAM65C
|
family with sequence similarity 65, member C |
| chr5_-_36301984 | 0.14 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr14_+_74034310 | 0.14 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr4_-_69111401 | 0.14 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr7_+_6655225 | 0.14 |
ENST00000457543.3
|
ZNF853
|
zinc finger protein 853 |
| chr6_+_13272904 | 0.14 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr15_+_43985725 | 0.13 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr21_+_17443434 | 0.13 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_45241109 | 0.13 |
ENST00000396651.3
|
RPS8
|
ribosomal protein S8 |
| chr12_+_15699286 | 0.13 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_-_65407524 | 0.13 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr19_+_36632204 | 0.13 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr15_+_80351977 | 0.13 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr20_+_62795827 | 0.12 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr6_+_150920999 | 0.12 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr10_-_21435488 | 0.12 |
ENST00000534331.1
ENST00000529198.1 ENST00000377118.4 |
C10orf113
|
chromosome 10 open reading frame 113 |
| chr14_-_69261310 | 0.12 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr14_+_39944025 | 0.12 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr7_-_73038867 | 0.12 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr2_-_50574856 | 0.12 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr11_+_125034586 | 0.12 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr8_-_7243080 | 0.12 |
ENST00000400156.4
|
ZNF705G
|
zinc finger protein 705G |
| chr7_-_73038822 | 0.12 |
ENST00000414749.2
ENST00000429400.2 ENST00000434326.1 |
MLXIPL
|
MLX interacting protein-like |
| chrX_+_7137475 | 0.12 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr1_-_68698222 | 0.12 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr9_-_28670283 | 0.12 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_-_50643664 | 0.11 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr18_-_53019208 | 0.11 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr8_-_37457350 | 0.11 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr14_-_47351391 | 0.11 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr1_-_10532531 | 0.11 |
ENST00000377036.2
ENST00000377038.3 |
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr3_-_112127981 | 0.11 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr16_-_85617170 | 0.11 |
ENST00000602862.1
|
RP11-118F19.1
|
RP11-118F19.1 |
| chr10_+_114710425 | 0.11 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_-_103749205 | 0.11 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_119302508 | 0.11 |
ENST00000442245.4
|
EMX2
|
empty spiracles homeobox 2 |
| chr3_-_98235962 | 0.11 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr2_+_196313239 | 0.10 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr12_+_56325812 | 0.10 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr20_-_31124186 | 0.10 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr8_+_22844995 | 0.10 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr1_-_2458026 | 0.10 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr9_+_125132803 | 0.10 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.7 | GO:0019082 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.2 | 1.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.2 | 1.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.6 | GO:0048631 | negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.4 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.1 | 0.4 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 1.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.5 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0051795 | catagen(GO:0042637) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.9 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.7 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.4 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.6 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.4 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.7 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 1.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:0061349 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.0 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.7 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.2 | 1.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.7 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.5 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.3 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0015254 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 1.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:1902379 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |