Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
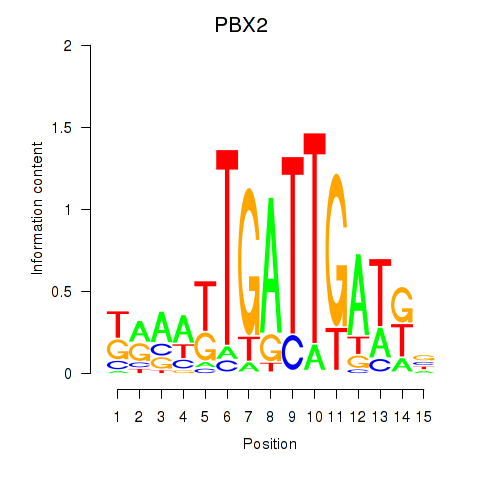
Results for PBX2
Z-value: 0.78
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX2 | hg19_v2_chr6_-_32157947_32157992 | -0.03 | 9.7e-01 | Click! |
Activity profile of PBX2 motif
Sorted Z-values of PBX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_1128781 | 0.51 |
ENST00000293897.4
ENST00000562758.1 |
SSTR5
|
somatostatin receptor 5 |
| chr14_-_55658323 | 0.47 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr9_-_128246769 | 0.41 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr1_-_26231589 | 0.36 |
ENST00000374291.1
|
STMN1
|
stathmin 1 |
| chrX_+_56590002 | 0.34 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr14_-_55658252 | 0.34 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr12_-_28124903 | 0.33 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr4_-_100065440 | 0.32 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr3_-_52864680 | 0.31 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr2_+_44396000 | 0.31 |
ENST00000409895.4
ENST00000409432.3 ENST00000282412.4 ENST00000378551.2 ENST00000345249.4 |
PPM1B
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr11_+_62432777 | 0.25 |
ENST00000532971.1
|
METTL12
|
methyltransferase like 12 |
| chr17_+_7758374 | 0.24 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr1_+_62417957 | 0.24 |
ENST00000307297.7
ENST00000543708.1 |
INADL
|
InaD-like (Drosophila) |
| chr4_+_71019903 | 0.22 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr11_-_111794446 | 0.21 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr7_+_134528635 | 0.21 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr12_+_53835508 | 0.20 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr14_-_54317532 | 0.19 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr2_+_20646824 | 0.19 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr7_-_32529973 | 0.19 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chrX_+_119737806 | 0.17 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr12_+_53835539 | 0.17 |
ENST00000547368.1
ENST00000379786.4 ENST00000551945.1 |
PRR13
|
proline rich 13 |
| chr22_+_42017987 | 0.16 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr2_-_99917639 | 0.15 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr5_-_39462390 | 0.14 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr14_+_58765305 | 0.13 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr11_+_68228186 | 0.12 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr4_+_71017791 | 0.12 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr1_+_101702417 | 0.11 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr12_+_53835383 | 0.11 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr12_-_120189900 | 0.10 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr12_-_39734783 | 0.09 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr4_-_100065419 | 0.08 |
ENST00000504125.1
ENST00000505590.1 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr10_-_45474237 | 0.08 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr2_+_234602305 | 0.08 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr3_+_129159039 | 0.07 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr2_-_31440377 | 0.07 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr9_-_20382446 | 0.07 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_-_39339777 | 0.06 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr2_+_27255806 | 0.06 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr3_+_129158926 | 0.06 |
ENST00000347300.2
ENST00000296266.3 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr12_-_28125638 | 0.06 |
ENST00000545234.1
|
PTHLH
|
parathyroid hormone-like hormone |
| chr16_-_11492366 | 0.05 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr22_-_30642728 | 0.05 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr1_+_56880606 | 0.05 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr6_+_160211481 | 0.05 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr7_-_112727774 | 0.04 |
ENST00000297146.3
ENST00000501255.2 |
GPR85
|
G protein-coupled receptor 85 |
| chr17_-_29151794 | 0.04 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr17_+_8191815 | 0.03 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr14_+_64854958 | 0.03 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr12_+_53835425 | 0.02 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr10_+_69869237 | 0.02 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr10_+_47894572 | 0.02 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr14_+_58765103 | 0.01 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr10_+_124320156 | 0.01 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr16_+_1832902 | 0.01 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr1_+_207038699 | 0.01 |
ENST00000367098.1
|
IL20
|
interleukin 20 |
| chr2_-_190044480 | 0.01 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 0.8 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.4 | GO:0070300 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |