Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
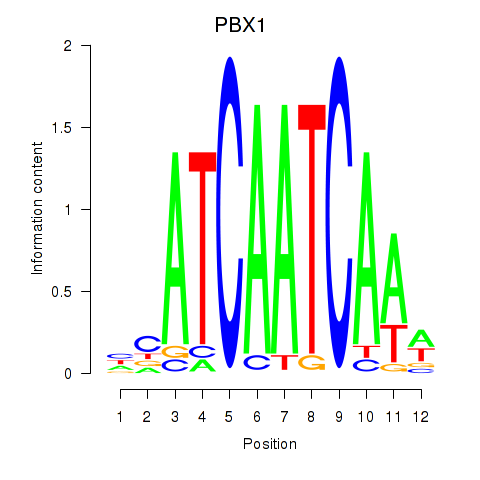
Results for PBX1
Z-value: 0.36
Transcription factors associated with PBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX1
|
ENSG00000185630.14 | PBX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PBX1 | hg19_v2_chr1_+_164528866_164528877 | -0.99 | 7.6e-03 | Click! |
Activity profile of PBX1 motif
Sorted Z-values of PBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_58018269 | 0.37 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr7_-_92747269 | 0.35 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chrX_-_1331527 | 0.29 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr22_+_50981079 | 0.25 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr21_-_16125773 | 0.24 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr16_+_15596123 | 0.24 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr17_+_7758374 | 0.23 |
ENST00000301599.6
ENST00000574668.1 |
TMEM88
|
transmembrane protein 88 |
| chr2_+_169926047 | 0.22 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr14_-_24584138 | 0.21 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chrX_+_1734051 | 0.20 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr6_-_32784687 | 0.19 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr6_+_27114861 | 0.18 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chrM_+_4431 | 0.17 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr5_-_94417186 | 0.17 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr15_-_60695071 | 0.16 |
ENST00000557904.1
|
ANXA2
|
annexin A2 |
| chr3_-_4927447 | 0.16 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chr22_-_30642728 | 0.16 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr15_-_60683326 | 0.16 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr7_-_7782204 | 0.16 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr21_+_44313375 | 0.15 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr2_+_12857015 | 0.15 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr22_-_30642782 | 0.15 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr16_-_88729473 | 0.15 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr6_+_151561085 | 0.15 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_-_26124138 | 0.14 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr6_-_26250835 | 0.14 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr5_-_141703713 | 0.13 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr11_-_95523500 | 0.13 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr1_+_156589198 | 0.13 |
ENST00000456112.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr5_-_94417314 | 0.13 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_28241226 | 0.12 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr10_+_24738355 | 0.12 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr11_+_44117219 | 0.12 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr10_-_18948208 | 0.12 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr7_-_559853 | 0.12 |
ENST00000405692.2
|
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr8_-_67874805 | 0.12 |
ENST00000563496.1
|
TCF24
|
transcription factor 24 |
| chr2_+_12857043 | 0.12 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr10_+_124320156 | 0.12 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_-_154335300 | 0.11 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr17_+_73663470 | 0.11 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr11_+_66384053 | 0.11 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
| chr16_+_19078960 | 0.11 |
ENST00000568985.1
ENST00000566110.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr2_+_201997595 | 0.11 |
ENST00000470178.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr6_+_151561506 | 0.11 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr19_-_5903714 | 0.11 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr17_-_26694979 | 0.11 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr19_+_41698927 | 0.11 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr17_+_73663402 | 0.11 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chr12_-_96389702 | 0.11 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr19_+_46498704 | 0.11 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr2_+_201997492 | 0.10 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_+_10828724 | 0.10 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr2_+_62132781 | 0.10 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr2_+_219110149 | 0.10 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chrX_-_140673133 | 0.10 |
ENST00000370519.3
|
SPANXA1
|
sperm protein associated with the nucleus, X-linked, family member A1 |
| chr5_-_94417339 | 0.09 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_-_26695013 | 0.09 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr14_+_23938891 | 0.09 |
ENST00000408901.3
ENST00000397154.3 ENST00000555128.1 |
NGDN
|
neuroguidin, EIF4E binding protein |
| chrM_+_14741 | 0.09 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr11_-_74800799 | 0.09 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr11_+_114166536 | 0.09 |
ENST00000299964.3
|
NNMT
|
nicotinamide N-methyltransferase |
| chr12_+_54402790 | 0.09 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr12_+_123944070 | 0.09 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr1_+_205473720 | 0.09 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr6_+_31553978 | 0.09 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr4_+_69962212 | 0.09 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr3_+_122785895 | 0.09 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr4_-_185776854 | 0.08 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr1_-_28241024 | 0.08 |
ENST00000313433.7
ENST00000444045.1 |
RPA2
|
replication protein A2, 32kDa |
| chr19_-_51141196 | 0.08 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr19_+_14640372 | 0.08 |
ENST00000215567.5
ENST00000598298.1 ENST00000596073.1 ENST00000600083.1 ENST00000436007.2 |
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr7_+_120629653 | 0.08 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_140677562 | 0.08 |
ENST00000370518.3
|
SPANXA2
|
SPANX family, member A2 |
| chrX_-_110655306 | 0.08 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr18_+_29598335 | 0.08 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr6_-_31782813 | 0.08 |
ENST00000375654.4
|
HSPA1L
|
heat shock 70kDa protein 1-like |
| chr17_+_75181292 | 0.08 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr2_+_201997676 | 0.08 |
ENST00000462763.1
ENST00000479953.2 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr19_-_39881669 | 0.08 |
ENST00000221266.7
|
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr6_-_167040731 | 0.08 |
ENST00000265678.4
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr2_+_62132800 | 0.08 |
ENST00000538736.1
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr16_-_29499154 | 0.08 |
ENST00000354563.5
|
RP11-231C14.4
|
Uncharacterized protein |
| chr2_-_200323414 | 0.08 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr16_+_19078911 | 0.08 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr4_-_72649763 | 0.08 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr6_-_15586238 | 0.07 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr6_+_36839616 | 0.07 |
ENST00000359359.2
ENST00000510325.2 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr5_-_177210399 | 0.07 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr5_-_171881362 | 0.07 |
ENST00000519643.1
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr11_+_68228186 | 0.07 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chrM_-_14670 | 0.07 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr10_+_5238793 | 0.07 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr10_+_69869237 | 0.07 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr22_+_35695793 | 0.07 |
ENST00000456128.1
ENST00000449058.2 ENST00000411850.1 ENST00000425375.1 ENST00000436462.2 ENST00000382034.5 |
TOM1
|
target of myb1 (chicken) |
| chr11_+_61717279 | 0.07 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chrX_-_92928557 | 0.07 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr19_-_49955050 | 0.07 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr6_-_51952418 | 0.07 |
ENST00000371117.3
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr19_-_39881777 | 0.07 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr16_+_19222479 | 0.07 |
ENST00000568433.1
|
SYT17
|
synaptotagmin XVII |
| chr19_+_5681153 | 0.07 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr10_+_124320195 | 0.07 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr2_-_158182322 | 0.07 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr12_-_109221160 | 0.07 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr16_+_69458537 | 0.07 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr16_+_55512742 | 0.07 |
ENST00000568715.1
ENST00000219070.4 |
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr19_-_58400148 | 0.06 |
ENST00000595048.1
ENST00000600634.1 ENST00000595295.1 ENST00000596604.1 ENST00000597342.1 ENST00000597807.1 |
ZNF814
|
zinc finger protein 814 |
| chr3_-_129158676 | 0.06 |
ENST00000393278.2
|
MBD4
|
methyl-CpG binding domain protein 4 |
| chr1_-_238108575 | 0.06 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr6_-_51952367 | 0.06 |
ENST00000340994.4
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr8_+_37887772 | 0.06 |
ENST00000338825.4
|
EIF4EBP1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr2_-_158182105 | 0.06 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr7_+_80275953 | 0.06 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_68797580 | 0.06 |
ENST00000539404.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr13_+_74805561 | 0.06 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr16_+_16484691 | 0.06 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr3_-_150264272 | 0.06 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr10_-_5638048 | 0.06 |
ENST00000478294.1
|
RP13-463N16.6
|
RP13-463N16.6 |
| chr4_-_184241927 | 0.06 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr8_-_36636676 | 0.06 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr21_-_16126181 | 0.06 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr10_+_50887683 | 0.05 |
ENST00000374113.3
ENST00000374111.3 ENST00000374112.3 ENST00000535836.1 |
C10orf53
|
chromosome 10 open reading frame 53 |
| chr4_+_26324474 | 0.05 |
ENST00000514675.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_-_63651939 | 0.05 |
ENST00000563855.1
ENST00000568932.1 ENST00000564290.1 |
RP11-368L12.1
|
RP11-368L12.1 |
| chr6_+_27775899 | 0.05 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr5_+_159656437 | 0.05 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr10_+_48189612 | 0.05 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr15_+_74421961 | 0.05 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr6_+_99282570 | 0.05 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr1_+_70687079 | 0.05 |
ENST00000395136.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr14_+_29236269 | 0.05 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr5_+_151151471 | 0.05 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr5_+_140213815 | 0.05 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr8_+_7801144 | 0.05 |
ENST00000443676.1
|
ZNF705B
|
zinc finger protein 705B |
| chr4_+_56815102 | 0.05 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr12_+_101988627 | 0.05 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr18_+_22040620 | 0.05 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr3_-_71353892 | 0.05 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr7_-_107883678 | 0.05 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr19_+_49999631 | 0.05 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr7_+_80275752 | 0.05 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr5_-_149535190 | 0.04 |
ENST00000517488.1
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr12_+_21525818 | 0.04 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr12_+_101988774 | 0.04 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr17_-_59668550 | 0.04 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr2_-_158182410 | 0.04 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr5_+_140207536 | 0.04 |
ENST00000529310.1
ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr17_-_15519008 | 0.04 |
ENST00000261644.8
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chrX_-_50386648 | 0.04 |
ENST00000460112.3
|
SHROOM4
|
shroom family member 4 |
| chrX_-_10645724 | 0.04 |
ENST00000413894.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr14_-_60097297 | 0.04 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr10_+_69865866 | 0.04 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr3_+_120626919 | 0.04 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chrX_+_150345054 | 0.04 |
ENST00000218316.3
|
GPR50
|
G protein-coupled receptor 50 |
| chr1_-_115323245 | 0.04 |
ENST00000060969.5
ENST00000369528.5 |
SIKE1
|
suppressor of IKBKE 1 |
| chr11_+_61717336 | 0.04 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr2_-_200322723 | 0.04 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr2_+_201242715 | 0.04 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr3_-_169587621 | 0.04 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr14_+_24583836 | 0.04 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr4_-_168155169 | 0.04 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr15_+_69222827 | 0.04 |
ENST00000310673.3
ENST00000448182.3 ENST00000260364.5 |
SPESP1
NOX5
|
sperm equatorial segment protein 1 NADPH oxidase, EF-hand calcium binding domain 5 |
| chr7_-_120498357 | 0.04 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr4_-_48014931 | 0.04 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr13_+_95254085 | 0.04 |
ENST00000376958.4
|
GPR180
|
G protein-coupled receptor 180 |
| chr3_-_143567262 | 0.04 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr12_-_133787772 | 0.04 |
ENST00000545350.1
|
AC226150.4
|
Uncharacterized protein |
| chr4_-_185776789 | 0.04 |
ENST00000510284.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr3_-_149510553 | 0.04 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr19_+_41770269 | 0.04 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_+_156589051 | 0.04 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chr12_+_48722763 | 0.04 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr19_-_3801789 | 0.04 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_+_35417939 | 0.04 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr19_-_49121054 | 0.04 |
ENST00000546623.1
ENST00000084795.5 |
RPL18
|
ribosomal protein L18 |
| chr15_+_69222909 | 0.04 |
ENST00000455873.3
|
NOX5
|
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr16_+_69221028 | 0.04 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr19_-_49955096 | 0.04 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr9_+_113431059 | 0.04 |
ENST00000416899.2
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase |
| chr9_-_32573130 | 0.03 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr5_+_125759140 | 0.03 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr10_-_105845674 | 0.03 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr3_+_159557637 | 0.03 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr17_+_45973516 | 0.03 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr17_+_46125707 | 0.03 |
ENST00000584137.1
ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chrX_+_140084756 | 0.03 |
ENST00000449283.1
|
SPANXB2
|
SPANX family, member B2 |
| chr14_-_69619689 | 0.03 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr3_-_53164423 | 0.03 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr19_-_14640005 | 0.03 |
ENST00000596853.1
ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr4_+_9212383 | 0.03 |
ENST00000421288.2
ENST00000417945.1 |
USP17L13
USP17L10
|
ubiquitin specific peptidase 17-like family member 13 ubiquitin specific peptidase 17-like family member 10 |
| chr18_-_48346415 | 0.03 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr8_+_99076750 | 0.03 |
ENST00000545282.1
|
C8orf47
|
chromosome 8 open reading frame 47 |
| chr4_+_69962185 | 0.03 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr2_+_27886330 | 0.03 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr2_-_39456673 | 0.03 |
ENST00000378803.1
ENST00000395035.3 |
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr1_+_104293028 | 0.03 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr11_+_34938119 | 0.03 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr5_-_10761206 | 0.03 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr8_-_42360015 | 0.03 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr9_-_101471479 | 0.03 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.3 | GO:0052229 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.2 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0035789 | cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.0 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |