Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
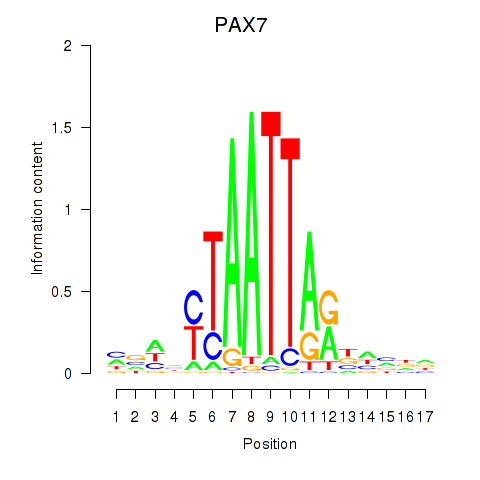
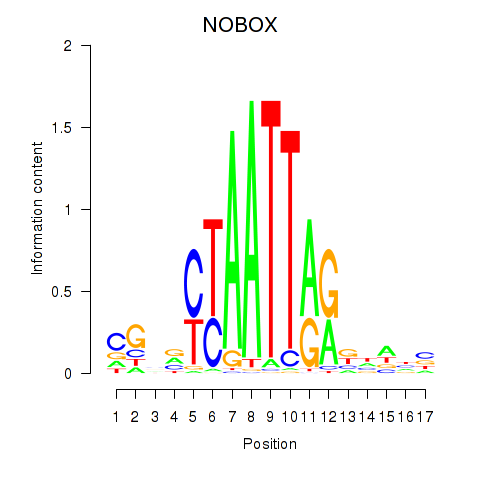
Results for PAX7_NOBOX
Z-value: 0.69
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX7
|
ENSG00000009709.7 | paired box 7 |
|
NOBOX
|
ENSG00000106410.10 | NOBOX oogenesis homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg19_v2_chr1_+_18957500_18957623 | -0.74 | 2.6e-01 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_33913708 | 0.96 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_-_121986923 | 0.81 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr11_-_102668879 | 0.77 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr10_+_91152303 | 0.67 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr19_-_3557570 | 0.66 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr22_-_32766972 | 0.63 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr5_+_53686658 | 0.62 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr17_-_19015945 | 0.61 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr12_+_113354341 | 0.57 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr7_+_37723336 | 0.54 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr17_+_19091325 | 0.48 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr11_-_327537 | 0.42 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr17_-_43045439 | 0.40 |
ENST00000253407.3
|
C1QL1
|
complement component 1, q subcomponent-like 1 |
| chr22_+_46476192 | 0.39 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr4_-_19458597 | 0.38 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr11_-_95523500 | 0.36 |
ENST00000540054.1
|
FAM76B
|
family with sequence similarity 76, member B |
| chr14_-_67878917 | 0.35 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chrX_+_10126488 | 0.35 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr2_+_232316906 | 0.34 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr15_+_80351910 | 0.34 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr7_-_105926058 | 0.33 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr6_+_24126350 | 0.33 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr3_-_108248169 | 0.33 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr11_-_74800799 | 0.33 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr10_+_90660832 | 0.32 |
ENST00000371924.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr6_+_34204642 | 0.30 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_+_182850551 | 0.30 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_-_225811747 | 0.30 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr11_-_62521614 | 0.29 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr1_-_190446759 | 0.28 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr19_-_54619006 | 0.28 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr12_+_26348246 | 0.28 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr6_+_55192267 | 0.28 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr10_-_17171817 | 0.26 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr7_-_87856303 | 0.26 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr6_-_31125850 | 0.25 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr17_-_39203519 | 0.23 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr12_-_31478428 | 0.23 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr19_-_40931891 | 0.23 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr22_-_32767017 | 0.22 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr16_-_29934558 | 0.21 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr13_+_19756173 | 0.21 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr1_-_183559693 | 0.21 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr17_+_56833184 | 0.21 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr4_+_169013666 | 0.21 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr12_-_22063787 | 0.21 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr4_-_185820602 | 0.21 |
ENST00000515864.1
ENST00000507183.1 |
RP11-701P16.4
|
long intergenic non-protein coding RNA 1093 |
| chr6_-_33256664 | 0.21 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr10_+_6821545 | 0.20 |
ENST00000436383.1
|
LINC00707
|
long intergenic non-protein coding RNA 707 |
| chr1_+_225600404 | 0.20 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr6_-_32806506 | 0.20 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr9_-_132383055 | 0.20 |
ENST00000372478.4
|
C9orf50
|
chromosome 9 open reading frame 50 |
| chr9_-_75488984 | 0.20 |
ENST00000423171.1
ENST00000449235.1 ENST00000453787.1 |
RP11-151D14.1
|
RP11-151D14.1 |
| chr6_+_26402465 | 0.20 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr21_-_27423339 | 0.19 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr3_+_47324424 | 0.19 |
ENST00000437353.1
ENST00000232766.5 ENST00000455924.2 |
KLHL18
|
kelch-like family member 18 |
| chr3_-_141747950 | 0.19 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_182850743 | 0.19 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr6_-_30684898 | 0.19 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr12_+_52695617 | 0.19 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr7_-_87856280 | 0.19 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr6_+_31553978 | 0.19 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr2_+_169926047 | 0.19 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr3_-_127317047 | 0.18 |
ENST00000462228.1
ENST00000490643.1 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr9_+_139780942 | 0.18 |
ENST00000247668.2
ENST00000359662.3 |
TRAF2
|
TNF receptor-associated factor 2 |
| chr1_+_201159914 | 0.18 |
ENST00000335211.4
ENST00000451870.2 ENST00000295591.8 |
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr17_+_50939459 | 0.18 |
ENST00000412360.1
|
AC102948.2
|
Uncharacterized protein |
| chr17_-_72772425 | 0.18 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_+_157963063 | 0.18 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr7_-_100183742 | 0.18 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr12_+_64798826 | 0.18 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr1_-_201140673 | 0.18 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr11_+_67798090 | 0.17 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_+_43855545 | 0.17 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr11_+_67798114 | 0.17 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chrX_-_48271344 | 0.17 |
ENST00000376884.2
ENST00000396928.1 |
SSX4B
|
synovial sarcoma, X breakpoint 4B |
| chr9_+_34652164 | 0.17 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr8_-_30584524 | 0.17 |
ENST00000521479.1
|
GSR
|
glutathione reductase |
| chr3_-_126327398 | 0.17 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr7_+_100136811 | 0.16 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr8_+_39972170 | 0.16 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr20_+_5731083 | 0.16 |
ENST00000445603.1
ENST00000442185.1 |
C20orf196
|
chromosome 20 open reading frame 196 |
| chr1_+_155278539 | 0.16 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr2_-_20551983 | 0.16 |
ENST00000442400.1
|
PUM2
|
pumilio RNA-binding family member 2 |
| chr10_-_25305011 | 0.16 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr1_+_155278625 | 0.16 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr20_-_56265680 | 0.16 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_+_100209979 | 0.16 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr7_+_100210133 | 0.16 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr10_-_99205607 | 0.16 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr17_-_72772462 | 0.15 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr1_+_12976450 | 0.15 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr19_-_54618650 | 0.15 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr1_+_151253991 | 0.15 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr2_+_234826016 | 0.15 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_+_72772621 | 0.15 |
ENST00000335464.5
ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104
|
transmembrane protein 104 |
| chr10_-_28571015 | 0.15 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_-_87849340 | 0.15 |
ENST00000419179.1
ENST00000265729.2 |
SRI
|
sorcin |
| chr1_-_183560011 | 0.14 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr12_-_25348007 | 0.14 |
ENST00000354189.5
ENST00000545133.1 ENST00000554347.1 ENST00000395987.3 ENST00000320267.9 ENST00000395990.2 ENST00000537577.1 |
CASC1
|
cancer susceptibility candidate 1 |
| chr19_-_6424783 | 0.14 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
| chr19_-_54663473 | 0.14 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr18_+_616672 | 0.14 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr16_-_66584059 | 0.14 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr6_+_43612750 | 0.14 |
ENST00000372165.4
ENST00000372163.4 |
RSPH9
|
radial spoke head 9 homolog (Chlamydomonas) |
| chr5_+_142149932 | 0.14 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chrX_-_153363125 | 0.14 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr10_+_103986085 | 0.14 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr6_+_170151708 | 0.14 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr22_-_30722912 | 0.14 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr15_+_62853562 | 0.14 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr15_+_89631647 | 0.14 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr16_-_66583994 | 0.14 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr19_+_19303720 | 0.13 |
ENST00000392324.4
|
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr18_-_61329118 | 0.13 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr11_-_65149422 | 0.13 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr13_+_78315348 | 0.13 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr5_+_115358088 | 0.13 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr3_+_63638372 | 0.13 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr12_-_54652060 | 0.13 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr15_+_66679155 | 0.13 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr9_-_38424443 | 0.13 |
ENST00000377694.1
|
IGFBPL1
|
insulin-like growth factor binding protein-like 1 |
| chr6_+_127898312 | 0.13 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr6_-_26043885 | 0.13 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr16_+_31724618 | 0.13 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr17_+_61473104 | 0.13 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_+_100209725 | 0.13 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr7_+_93535866 | 0.13 |
ENST00000429473.1
ENST00000430875.1 ENST00000428834.1 |
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_-_60683326 | 0.13 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr7_-_99698338 | 0.13 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr2_-_87248975 | 0.13 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr17_-_76732928 | 0.12 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr4_-_113558079 | 0.12 |
ENST00000445203.2
|
C4orf21
|
chromosome 4 open reading frame 21 |
| chr6_-_26124138 | 0.12 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr19_+_19303572 | 0.12 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr12_+_38710555 | 0.12 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr12_-_118628315 | 0.12 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr13_-_30160925 | 0.12 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_59383617 | 0.12 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr6_-_32145861 | 0.12 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr14_+_32798462 | 0.11 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_-_190927447 | 0.11 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr2_-_198540751 | 0.11 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr19_+_4153598 | 0.11 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr2_+_10091783 | 0.11 |
ENST00000324883.5
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr18_+_39535239 | 0.11 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr11_+_67798363 | 0.11 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_-_13390765 | 0.11 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr17_+_74536164 | 0.11 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr12_-_10022735 | 0.11 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr10_+_5135981 | 0.11 |
ENST00000380554.3
|
AKR1C3
|
aldo-keto reductase family 1, member C3 |
| chr15_+_69745123 | 0.11 |
ENST00000260379.6
ENST00000357790.5 ENST00000560274.1 |
RPLP1
|
ribosomal protein, large, P1 |
| chr2_-_96700664 | 0.11 |
ENST00000359548.4
ENST00000377137.3 ENST00000439254.1 ENST00000453542.1 |
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_-_74618964 | 0.11 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr8_-_62559366 | 0.11 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr17_+_79650962 | 0.11 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr8_-_82024290 | 0.11 |
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr17_+_46184911 | 0.11 |
ENST00000580219.1
ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11
|
sorting nexin 11 |
| chr7_+_116654958 | 0.11 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr7_-_150020578 | 0.11 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr4_-_74486347 | 0.10 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr15_+_64680003 | 0.10 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr3_+_52245458 | 0.10 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr2_+_145780725 | 0.10 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr19_+_19303008 | 0.10 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr1_+_31769836 | 0.10 |
ENST00000344147.5
ENST00000373714.1 ENST00000546109.1 ENST00000422613.2 |
ZCCHC17
|
zinc finger, CCHC domain containing 17 |
| chr20_+_54967409 | 0.10 |
ENST00000415828.1
ENST00000217109.4 ENST00000428552.1 |
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa |
| chr4_+_158493642 | 0.10 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr12_+_34175398 | 0.10 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr14_-_20929624 | 0.10 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr1_-_224624730 | 0.10 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr14_-_24711470 | 0.10 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_44115814 | 0.10 |
ENST00000372396.3
|
KDM4A
|
lysine (K)-specific demethylase 4A |
| chr3_+_127317705 | 0.10 |
ENST00000480910.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr2_+_143635067 | 0.10 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chr4_-_72649763 | 0.10 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr2_-_74619152 | 0.10 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr6_+_3259148 | 0.10 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr4_+_74347400 | 0.10 |
ENST00000226355.3
|
AFM
|
afamin |
| chrX_-_92928557 | 0.10 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr11_-_128894053 | 0.10 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr10_-_112064665 | 0.10 |
ENST00000369603.5
|
SMNDC1
|
survival motor neuron domain containing 1 |
| chr19_+_18699535 | 0.09 |
ENST00000358607.6
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chrX_-_48815633 | 0.09 |
ENST00000428668.2
|
OTUD5
|
OTU domain containing 5 |
| chr11_+_62529008 | 0.09 |
ENST00000301788.7
ENST00000533442.1 |
POLR2G
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr11_-_108093329 | 0.09 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr20_+_55967129 | 0.09 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr2_+_183982255 | 0.09 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr17_+_74536115 | 0.09 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr11_-_35287243 | 0.09 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_+_97709725 | 0.09 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr14_-_24711806 | 0.09 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr10_+_70980051 | 0.09 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr15_+_58430567 | 0.09 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr3_-_52719546 | 0.09 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr8_-_135522425 | 0.09 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr5_+_102200948 | 0.09 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr19_+_18699599 | 0.09 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr4_-_70518941 | 0.09 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) negative regulation of heart rate(GO:0010459) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.3 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.3 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 0.3 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0052229 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0060488 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0060928 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.3 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.1 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.2 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0015265 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |