Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
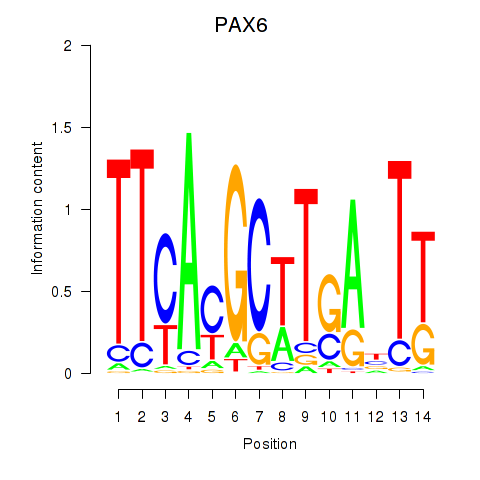
Results for PAX6
Z-value: 1.27
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg19_v2_chr11_-_31839488_31839515 | 0.91 | 9.4e-02 | Click! |
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_121476959 | 1.75 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr1_+_79115503 | 1.58 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr12_+_113354341 | 1.53 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_+_91092241 | 1.44 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr12_-_10007448 | 0.89 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr12_-_78934441 | 0.87 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chr19_-_36019123 | 0.61 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr1_+_223354486 | 0.59 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr8_-_19614810 | 0.57 |
ENST00000524213.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr20_+_48807351 | 0.55 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr3_+_108015382 | 0.53 |
ENST00000463019.3
ENST00000491820.1 ENST00000467562.1 ENST00000482430.1 ENST00000462629.1 |
HHLA2
|
HERV-H LTR-associating 2 |
| chr22_-_30968813 | 0.52 |
ENST00000443111.2
ENST00000443136.1 ENST00000426220.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chrX_-_124097620 | 0.49 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr19_-_37701386 | 0.46 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr22_-_30968839 | 0.45 |
ENST00000445645.1
ENST00000416358.1 ENST00000423371.1 ENST00000411821.1 ENST00000448604.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr18_-_21242774 | 0.45 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr4_-_129491686 | 0.44 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr2_-_86116020 | 0.44 |
ENST00000525834.2
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr16_+_77233294 | 0.43 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr12_+_113416340 | 0.42 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr18_+_61445205 | 0.41 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chrX_+_10126488 | 0.41 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr19_+_35939154 | 0.41 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr12_+_10460417 | 0.40 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr12_-_3982511 | 0.40 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr5_+_67485704 | 0.40 |
ENST00000520762.1
|
RP11-404L6.2
|
Uncharacterized protein |
| chr18_-_21242833 | 0.39 |
ENST00000586087.1
ENST00000592179.1 |
ANKRD29
|
ankyrin repeat domain 29 |
| chr21_+_39628655 | 0.39 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr4_+_130017268 | 0.37 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr14_-_36645674 | 0.37 |
ENST00000556013.2
|
PTCSC3
|
papillary thyroid carcinoma susceptibility candidate 3 (non-protein coding) |
| chr9_-_140142222 | 0.35 |
ENST00000344774.4
ENST00000388932.2 |
FAM166A
|
family with sequence similarity 166, member A |
| chr4_+_129349188 | 0.34 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr20_+_32250079 | 0.34 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr1_+_207818460 | 0.34 |
ENST00000508064.2
|
CR1L
|
complement component (3b/4b) receptor 1-like |
| chr4_+_123747979 | 0.32 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr4_+_70916119 | 0.32 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr4_+_70894130 | 0.31 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_-_110933663 | 0.30 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr1_-_2126192 | 0.30 |
ENST00000378546.4
|
C1orf86
|
chromosome 1 open reading frame 86 |
| chr8_-_18711866 | 0.30 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr22_+_39745930 | 0.30 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr9_+_127023704 | 0.30 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr12_+_83080724 | 0.29 |
ENST00000548305.1
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr12_+_93115281 | 0.29 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr3_+_140981456 | 0.29 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr12_-_3982548 | 0.28 |
ENST00000397096.2
ENST00000447133.3 ENST00000450737.2 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr11_+_74811578 | 0.28 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr3_-_119182506 | 0.27 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chrX_-_80457385 | 0.27 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr9_-_16870704 | 0.27 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr22_-_36220420 | 0.26 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_-_110933611 | 0.26 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chrX_+_106449862 | 0.26 |
ENST00000372453.3
ENST00000535523.1 |
PIH1D3
|
PIH1 domain containing 3 |
| chr3_-_171489085 | 0.26 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr1_-_232598163 | 0.26 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_-_10047219 | 0.26 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chr2_+_149402989 | 0.26 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr3_+_171561127 | 0.26 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr6_+_42896865 | 0.25 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr13_+_53602894 | 0.25 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr5_-_159766528 | 0.25 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr12_-_122884553 | 0.25 |
ENST00000535290.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr12_-_69080590 | 0.25 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr7_-_91808845 | 0.25 |
ENST00000343318.5
|
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr18_+_61445007 | 0.25 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_+_78224667 | 0.25 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr2_-_158182322 | 0.25 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_-_26033796 | 0.24 |
ENST00000259791.2
|
HIST1H2AB
|
histone cluster 1, H2ab |
| chr2_-_158182105 | 0.24 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr4_-_10686475 | 0.24 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr12_-_85306594 | 0.23 |
ENST00000266682.5
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr1_+_244227632 | 0.23 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr3_-_124839648 | 0.23 |
ENST00000430155.2
|
SLC12A8
|
solute carrier family 12, member 8 |
| chr4_-_174320687 | 0.23 |
ENST00000296506.3
|
SCRG1
|
stimulator of chondrogenesis 1 |
| chr17_+_36283971 | 0.23 |
ENST00000327454.6
ENST00000378174.5 |
TBC1D3F
|
TBC1 domain family, member 3F |
| chr6_-_30684898 | 0.22 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr17_+_25958174 | 0.22 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr2_+_114195268 | 0.22 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr7_+_76109827 | 0.22 |
ENST00000446820.2
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr22_-_30642728 | 0.22 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr12_-_70093162 | 0.22 |
ENST00000551160.1
|
BEST3
|
bestrophin 3 |
| chr14_+_57671888 | 0.21 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr3_+_151531810 | 0.21 |
ENST00000232892.7
|
AADAC
|
arylacetamide deacetylase |
| chr5_+_96212185 | 0.21 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr2_-_86116093 | 0.20 |
ENST00000377332.3
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr1_+_180897269 | 0.20 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr2_-_153573965 | 0.20 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr2_+_179184955 | 0.19 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein-like 6 |
| chr3_+_15045419 | 0.19 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr18_+_61420169 | 0.19 |
ENST00000425392.1
ENST00000336429.2 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr15_+_51973708 | 0.19 |
ENST00000558709.1
|
SCG3
|
secretogranin III |
| chr6_+_170151708 | 0.19 |
ENST00000592367.1
ENST00000590711.1 ENST00000366772.2 ENST00000592745.1 ENST00000392095.4 ENST00000366773.3 ENST00000586341.1 ENST00000418781.3 ENST00000588437.1 |
ERMARD
|
ER membrane-associated RNA degradation |
| chr6_+_26156551 | 0.18 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chrX_-_53461305 | 0.17 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr3_-_111852061 | 0.17 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr2_+_196522032 | 0.17 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr12_+_56915713 | 0.17 |
ENST00000262031.5
ENST00000552247.2 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr7_-_43909090 | 0.17 |
ENST00000317534.5
|
MRPS24
|
mitochondrial ribosomal protein S24 |
| chr8_+_102064237 | 0.17 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1 |
| chr3_-_87842631 | 0.16 |
ENST00000462792.1
|
RP11-451B8.1
|
RP11-451B8.1 |
| chr8_-_19615382 | 0.16 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr17_+_16945820 | 0.16 |
ENST00000577514.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr11_-_3078838 | 0.16 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr2_+_228189867 | 0.16 |
ENST00000423098.1
ENST00000304593.9 |
MFF
|
mitochondrial fission factor |
| chr8_-_62602327 | 0.16 |
ENST00000445642.3
ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH
|
aspartate beta-hydroxylase |
| chr17_-_73389737 | 0.16 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr3_+_151531859 | 0.16 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chrX_-_138914394 | 0.16 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr7_-_150754935 | 0.16 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr16_+_72088376 | 0.16 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr8_-_19615538 | 0.15 |
ENST00000517494.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_-_162931052 | 0.15 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr12_-_70093235 | 0.15 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr1_-_87379785 | 0.15 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr2_+_119699864 | 0.15 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr1_-_2126174 | 0.15 |
ENST00000400919.3
ENST00000420515.1 ENST00000378543.2 ENST00000400918.3 |
C1orf86
|
chromosome 1 open reading frame 86 |
| chr11_+_14926543 | 0.15 |
ENST00000523376.1
|
CALCB
|
calcitonin-related polypeptide beta |
| chr3_-_111852128 | 0.14 |
ENST00000308910.4
|
GCSAM
|
germinal center-associated, signaling and motility |
| chr5_+_53686658 | 0.14 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr6_-_134373732 | 0.14 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr1_-_85725316 | 0.14 |
ENST00000344356.5
ENST00000471115.1 |
C1orf52
|
chromosome 1 open reading frame 52 |
| chr12_-_15103621 | 0.14 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_+_168675182 | 0.14 |
ENST00000305861.1
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr17_-_77023683 | 0.14 |
ENST00000581579.1
|
C1QTNF1-AS1
|
C1QTNF1 antisense RNA 1 |
| chr17_-_73266616 | 0.14 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr20_+_9966728 | 0.14 |
ENST00000449270.1
|
RP5-839B4.8
|
RP5-839B4.8 |
| chr5_-_114938090 | 0.13 |
ENST00000427199.2
|
TICAM2
|
toll-like receptor adaptor molecule 2 |
| chr15_+_68924327 | 0.13 |
ENST00000543950.1
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr12_+_47473369 | 0.13 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr11_-_102401469 | 0.13 |
ENST00000260227.4
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr9_+_131549483 | 0.12 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr17_-_58096336 | 0.12 |
ENST00000587125.1
ENST00000407042.3 |
TBC1D3P1-DHX40P1
|
TBC1D3P1-DHX40P1 readthrough transcribed pseudogene |
| chr12_+_83080659 | 0.12 |
ENST00000321196.3
|
TMTC2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr4_-_88141615 | 0.12 |
ENST00000545252.1
ENST00000425278.2 ENST00000498875.2 |
KLHL8
|
kelch-like family member 8 |
| chr2_+_238877424 | 0.12 |
ENST00000434655.1
|
UBE2F
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr6_-_43027105 | 0.12 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr1_+_39491984 | 0.12 |
ENST00000372969.3
ENST00000372967.3 |
NDUFS5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) |
| chr5_-_58882219 | 0.12 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_50936142 | 0.12 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr14_+_29241910 | 0.12 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr1_-_23504176 | 0.12 |
ENST00000302291.4
|
LUZP1
|
leucine zipper protein 1 |
| chr15_-_78358465 | 0.12 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr22_-_37571089 | 0.11 |
ENST00000453962.1
ENST00000429622.1 ENST00000445595.1 |
IL2RB
|
interleukin 2 receptor, beta |
| chr7_-_150864635 | 0.11 |
ENST00000297537.4
|
GBX1
|
gastrulation brain homeobox 1 |
| chr8_+_41347915 | 0.11 |
ENST00000518270.1
ENST00000520817.1 |
GOLGA7
|
golgin A7 |
| chr1_+_218683438 | 0.11 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr6_+_143447322 | 0.11 |
ENST00000458219.1
|
AIG1
|
androgen-induced 1 |
| chr2_-_677369 | 0.11 |
ENST00000281017.3
|
TMEM18
|
transmembrane protein 18 |
| chr11_+_33563618 | 0.11 |
ENST00000526400.1
|
KIAA1549L
|
KIAA1549-like |
| chr2_-_158182410 | 0.11 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr16_+_77822427 | 0.11 |
ENST00000302536.2
|
VAT1L
|
vesicle amine transport 1-like |
| chr8_-_59412717 | 0.11 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr2_-_154335300 | 0.11 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr10_-_104597286 | 0.11 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr1_+_15668240 | 0.11 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr6_-_88299678 | 0.11 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr12_+_56915776 | 0.10 |
ENST00000550726.1
ENST00000542360.1 |
RBMS2
|
RNA binding motif, single stranded interacting protein 2 |
| chr7_-_138348969 | 0.10 |
ENST00000436657.1
|
SVOPL
|
SVOP-like |
| chr4_+_123747834 | 0.10 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr2_+_29320571 | 0.10 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr5_-_55529115 | 0.10 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chrX_-_92928557 | 0.10 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr6_-_26043885 | 0.10 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr2_+_202098166 | 0.10 |
ENST00000392263.2
ENST00000264274.9 ENST00000392259.2 ENST00000392266.3 ENST00000432109.2 ENST00000264275.5 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr4_+_40751914 | 0.10 |
ENST00000381782.2
ENST00000316607.5 |
NSUN7
|
NOP2/Sun domain family, member 7 |
| chr16_+_89228757 | 0.10 |
ENST00000565008.1
|
LINC00304
|
long intergenic non-protein coding RNA 304 |
| chr18_+_13611431 | 0.10 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_+_131549610 | 0.10 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr5_+_322733 | 0.10 |
ENST00000515206.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr15_+_51973680 | 0.10 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr12_+_111284764 | 0.10 |
ENST00000545036.1
ENST00000308208.5 |
CCDC63
|
coiled-coil domain containing 63 |
| chr4_+_40194609 | 0.10 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr12_-_106697974 | 0.10 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr7_+_90338712 | 0.09 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr19_-_50370509 | 0.09 |
ENST00000596014.1
|
PNKP
|
polynucleotide kinase 3'-phosphatase |
| chr18_+_18822185 | 0.09 |
ENST00000424526.1
ENST00000400483.4 ENST00000431264.1 |
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr15_+_42565393 | 0.09 |
ENST00000561871.1
|
GANC
|
glucosidase, alpha; neutral C |
| chr7_-_76255444 | 0.09 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr21_+_39628780 | 0.09 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr12_-_85306562 | 0.09 |
ENST00000551612.1
ENST00000450363.3 ENST00000552192.1 |
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr15_-_23034322 | 0.09 |
ENST00000539711.2
ENST00000560039.1 ENST00000398013.3 ENST00000337451.3 ENST00000359727.4 ENST00000398014.2 |
NIPA2
|
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr11_+_65554493 | 0.09 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr4_+_115519577 | 0.09 |
ENST00000310836.6
|
UGT8
|
UDP glycosyltransferase 8 |
| chr1_+_203651937 | 0.09 |
ENST00000341360.2
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr5_-_98262240 | 0.09 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr9_+_87283430 | 0.09 |
ENST00000376214.1
ENST00000376213.1 |
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr3_-_157251383 | 0.09 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr19_+_51628165 | 0.08 |
ENST00000250360.3
ENST00000440804.3 |
SIGLEC9
|
sialic acid binding Ig-like lectin 9 |
| chr13_-_24007815 | 0.08 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr17_+_72270429 | 0.08 |
ENST00000311014.6
|
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr15_-_65282274 | 0.08 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr2_+_202098203 | 0.08 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr2_-_74776586 | 0.08 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_+_128563652 | 0.08 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_+_13611763 | 0.08 |
ENST00000585931.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr19_-_41903161 | 0.08 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chrX_-_15332665 | 0.08 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr12_+_59194154 | 0.08 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr6_-_131291572 | 0.08 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_+_113970772 | 0.07 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.1 | 0.4 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.9 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 1.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.4 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.2 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.2 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.6 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.3 | GO:0015820 | leucine transport(GO:0015820) proline transmembrane transport(GO:0035524) |
| 0.0 | 3.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.4 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 0.6 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.9 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 1.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.4 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.2 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0030548 | ErbB-2 class receptor binding(GO:0005176) acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |