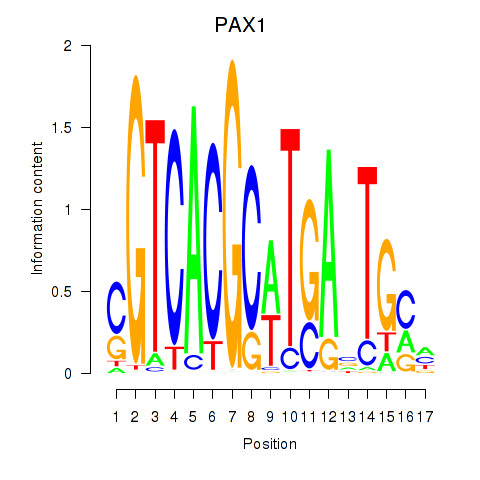
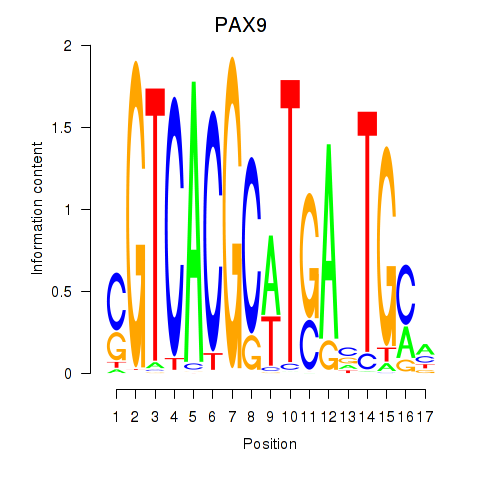
|
chr8_-_23261589
Show fit
|
0.32 |
ENST00000524168.1
ENST00000523833.2
ENST00000519243.1
ENST00000389131.3
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr8_-_23282797
Show fit
|
0.25 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr8_-_23282820
Show fit
|
0.23 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_+_75230853
Show fit
|
0.17 |
ENST00000244869.2
|
EREG
|
epiregulin
|
|
chr18_-_47813940
Show fit
|
0.16 |
ENST00000586837.1
ENST00000412036.2
ENST00000589940.1
|
CXXC1
|
CXXC finger protein 1
|
|
chr18_-_47814032
Show fit
|
0.15 |
ENST00000589548.1
ENST00000591474.1
|
CXXC1
|
CXXC finger protein 1
|
|
chrX_-_153707545
Show fit
|
0.11 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3
|
|
chr2_+_119699864
Show fit
|
0.10 |
ENST00000541757.1
ENST00000412481.1
|
MARCO
|
macrophage receptor with collagenous structure
|
|
chrX_-_153707246
Show fit
|
0.08 |
ENST00000407062.1
|
LAGE3
|
L antigen family, member 3
|
|
chr20_-_17641097
Show fit
|
0.07 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1
|
|
chr11_-_67397371
Show fit
|
0.06 |
ENST00000376693.2
ENST00000301490.4
|
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8
|
|
chr21_-_33104367
Show fit
|
0.05 |
ENST00000286835.7
ENST00000399804.1
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr4_+_123747979
Show fit
|
0.05 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr19_-_39104556
Show fit
|
0.04 |
ENST00000423454.2
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr7_-_2349579
Show fit
|
0.04 |
ENST00000435060.1
|
SNX8
|
sorting nexin 8
|
|
chr7_-_19157248
Show fit
|
0.04 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1
|
|
chr19_-_40791211
Show fit
|
0.04 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr19_-_409134
Show fit
|
0.04 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr5_-_133747551
Show fit
|
0.04 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like
|
|
chr18_-_33077556
Show fit
|
0.03 |
ENST00000589273.1
ENST00000586489.1
|
INO80C
|
INO80 complex subunit C
|
|
chr16_+_67197288
Show fit
|
0.03 |
ENST00000264009.8
ENST00000421453.1
|
HSF4
|
heat shock transcription factor 4
|
|
chr6_-_137366163
Show fit
|
0.03 |
ENST00000367748.1
|
IL20RA
|
interleukin 20 receptor, alpha
|
|
chr2_+_234160217
Show fit
|
0.03 |
ENST00000392017.4
ENST00000347464.5
ENST00000444735.1
ENST00000373525.5
ENST00000419681.1
|
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae)
|
|
chr7_-_22234381
Show fit
|
0.03 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr2_+_234160340
Show fit
|
0.02 |
ENST00000417017.1
ENST00000392020.4
ENST00000392018.1
|
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae)
|
|
chr17_-_7137582
Show fit
|
0.02 |
ENST00000575756.1
ENST00000575458.1
|
DVL2
|
dishevelled segment polarity protein 2
|
|
chr7_-_144533074
Show fit
|
0.02 |
ENST00000360057.3
ENST00000378099.3
|
TPK1
|
thiamin pyrophosphokinase 1
|
|
chr19_-_5838768
Show fit
|
0.02 |
ENST00000527106.1
ENST00000531199.1
ENST00000529165.1
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr17_+_37356528
Show fit
|
0.02 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19
|
|
chr11_-_126138808
Show fit
|
0.02 |
ENST00000332118.6
ENST00000532259.1
|
SRPR
|
signal recognition particle receptor (docking protein)
|
|
chr8_+_102064237
Show fit
|
0.02 |
ENST00000514926.1
|
RP11-302J23.1
|
RP11-302J23.1
|
|
chr11_+_12308447
Show fit
|
0.02 |
ENST00000256186.2
|
MICALCL
|
MICAL C-terminal like
|
|
chr14_-_69261310
Show fit
|
0.02 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1
|
|
chr18_-_47018769
Show fit
|
0.02 |
ENST00000583637.1
ENST00000578528.1
ENST00000578532.1
ENST00000580387.1
ENST00000579248.1
ENST00000581373.1
|
RPL17
|
ribosomal protein L17
|
|
chr11_+_6947720
Show fit
|
0.02 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215
|
|
chr17_+_76374714
Show fit
|
0.02 |
ENST00000262764.6
ENST00000589689.1
ENST00000329897.7
ENST00000592043.1
ENST00000587356.1
|
PGS1
|
phosphatidylglycerophosphate synthase 1
|
|
chr10_-_99393242
Show fit
|
0.02 |
ENST00000370635.3
ENST00000478953.1
ENST00000335628.3
|
MORN4
|
MORN repeat containing 4
|
|
chr6_+_159291090
Show fit
|
0.02 |
ENST00000367073.4
ENST00000608817.1
|
C6orf99
|
chromosome 6 open reading frame 99
|
|
chr17_+_37356586
Show fit
|
0.02 |
ENST00000579260.1
ENST00000582193.1
|
RPL19
|
ribosomal protein L19
|
|
chr17_+_37356555
Show fit
|
0.02 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19
|
|
chr17_-_39968406
Show fit
|
0.01 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4
|
|
chr18_-_47017956
Show fit
|
0.01 |
ENST00000584895.1
ENST00000332968.6
ENST00000580210.1
ENST00000579408.1
|
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr2_-_175711978
Show fit
|
0.01 |
ENST00000409089.2
|
CHN1
|
chimerin 1
|
|
chr3_+_127771212
Show fit
|
0.01 |
ENST00000243253.3
ENST00000481210.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chrX_+_148622513
Show fit
|
0.01 |
ENST00000393985.3
ENST00000423421.1
ENST00000423540.2
ENST00000434353.2
ENST00000514208.1
|
CXorf40A
|
chromosome X open reading frame 40A
|
|
chr1_+_45140360
Show fit
|
0.01 |
ENST00000418644.1
ENST00000458657.2
ENST00000441519.1
ENST00000535358.1
ENST00000445071.1
|
C1orf228
|
chromosome 1 open reading frame 228
|
|
chr6_-_137494775
Show fit
|
0.01 |
ENST00000349184.4
ENST00000296980.2
ENST00000339602.3
|
IL22RA2
|
interleukin 22 receptor, alpha 2
|
|
chr10_+_82297658
Show fit
|
0.01 |
ENST00000339284.2
|
SH2D4B
|
SH2 domain containing 4B
|
|
chr6_+_74104471
Show fit
|
0.01 |
ENST00000370336.4
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
|
|
chr1_-_204165610
Show fit
|
0.01 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor
|
|
chr8_+_39792474
Show fit
|
0.01 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2
|
|
chr10_-_99393208
Show fit
|
0.01 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4
|
|
chr17_-_39968855
Show fit
|
0.01 |
ENST00000355468.3
ENST00000590496.1
|
LEPREL4
|
leprecan-like 4
|
|
chr22_+_39745930
Show fit
|
0.01 |
ENST00000318801.4
ENST00000216155.7
ENST00000406293.3
ENST00000328933.5
|
SYNGR1
|
synaptogyrin 1
|
|
chr12_-_101801505
Show fit
|
0.01 |
ENST00000539055.1
ENST00000551688.1
ENST00000551671.1
ENST00000261636.8
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chrX_-_132091284
Show fit
|
0.01 |
ENST00000370833.2
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr2_-_39414848
Show fit
|
0.01 |
ENST00000451199.1
|
CDKL4
|
cyclin-dependent kinase-like 4
|
|
chr3_-_57113281
Show fit
|
0.01 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr15_+_63569731
Show fit
|
0.01 |
ENST00000261879.5
|
APH1B
|
APH1B gamma secretase subunit
|
|
chr11_+_6947647
Show fit
|
0.01 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215
|
|
chr1_+_26758849
Show fit
|
0.00 |
ENST00000533087.1
ENST00000531312.1
ENST00000525165.1
ENST00000525326.1
ENST00000525546.1
ENST00000436153.2
ENST00000530781.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr15_+_63569785
Show fit
|
0.00 |
ENST00000380343.4
ENST00000560353.1
|
APH1B
|
APH1B gamma secretase subunit
|
|
chr1_+_26758790
Show fit
|
0.00 |
ENST00000427245.2
ENST00000525682.2
ENST00000236342.7
ENST00000526219.1
ENST00000374185.3
ENST00000360009.2
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr19_-_40791302
Show fit
|
0.00 |
ENST00000392038.2
ENST00000578123.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr9_+_6716478
Show fit
|
0.00 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3
|
|
chr6_+_3118926
Show fit
|
0.00 |
ENST00000380379.5
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase)
|
|
chr2_-_162931052
Show fit
|
0.00 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr14_+_64854958
Show fit
|
0.00 |
ENST00000555709.2
ENST00000554739.1
ENST00000554768.1
ENST00000216605.8
|
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase
|
|
chr16_-_84076241
Show fit
|
0.00 |
ENST00000568178.1
|
SLC38A8
|
solute carrier family 38, member 8
|
|
chr5_-_133747589
Show fit
|
0.00 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like
|
|
chr4_+_619347
Show fit
|
0.00 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta
|