Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
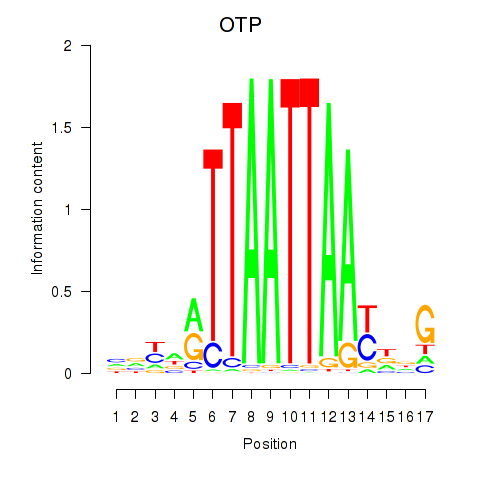
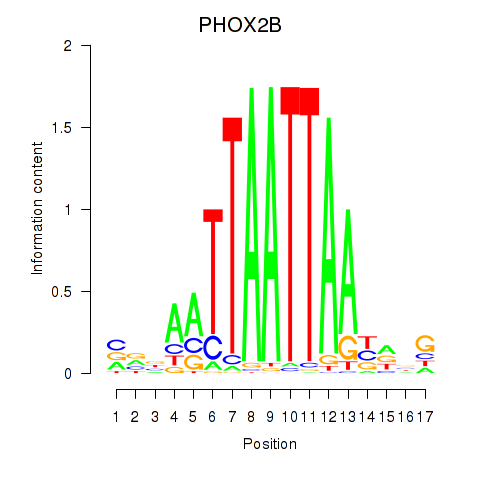
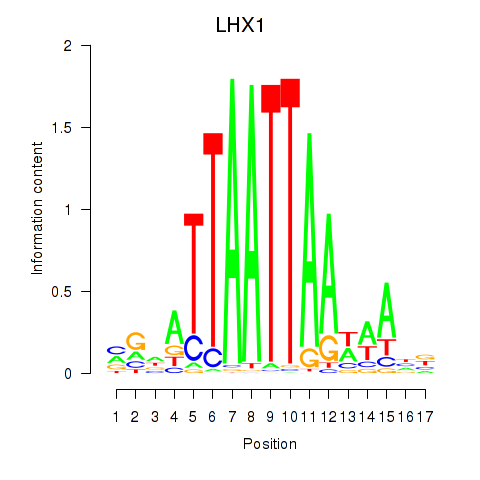
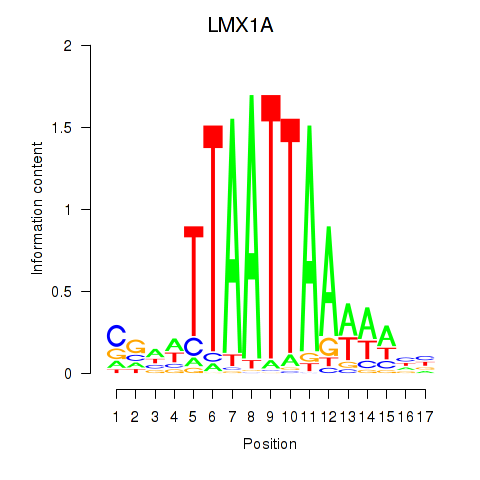
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 0.29
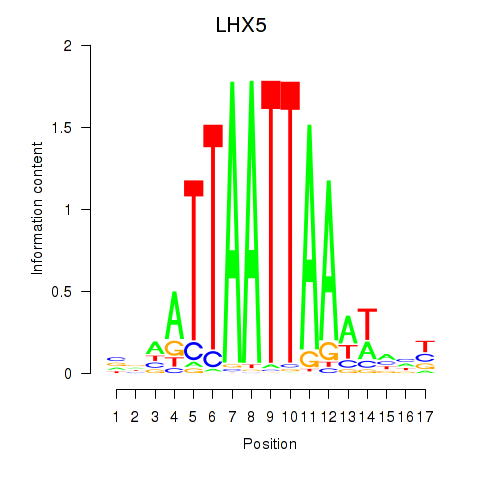
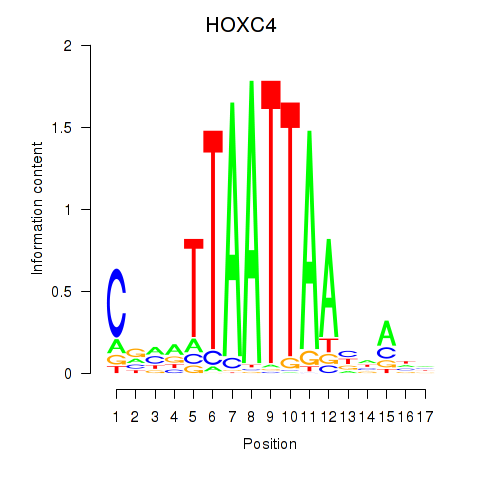
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.6 | orthopedia homeobox |
|
PHOX2B
|
ENSG00000109132.5 | paired like homeobox 2B |
|
LHX1
|
ENSG00000132130.7 | LIM homeobox 1 |
|
LMX1A
|
ENSG00000162761.10 | LIM homeobox transcription factor 1 alpha |
|
LHX5
|
ENSG00000089116.3 | LIM homeobox 5 |
|
HOXC4
|
ENSG00000198353.6 | homeobox C4 |
|
HOXC4
|
ENSG00000273266.1 | homeobox C4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC4 | hg19_v2_chr12_+_54447637_54447661 | -0.62 | 3.8e-01 | Click! |
| LHX5 | hg19_v2_chr12_-_113909877_113909877 | -0.29 | 7.1e-01 | Click! |
| OTP | hg19_v2_chr5_-_76935513_76935513 | 0.14 | 8.6e-01 | Click! |
| LHX1 | hg19_v2_chr17_+_35294075_35294102 | 0.00 | 1.0e+00 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_121986923 | 0.51 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr6_-_47445214 | 0.27 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr4_+_113568207 | 0.24 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_152178320 | 0.22 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr17_-_4938712 | 0.22 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr19_+_50016610 | 0.20 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_182850743 | 0.20 |
ENST00000409702.1
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_-_190446759 | 0.18 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr1_-_201140673 | 0.17 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr7_-_22862406 | 0.17 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr5_+_81575281 | 0.17 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr6_+_3259148 | 0.17 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr2_+_182850551 | 0.16 |
ENST00000452904.1
ENST00000409137.3 ENST00000280295.3 |
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_234826016 | 0.15 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_-_72772425 | 0.15 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr19_+_50016411 | 0.15 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_-_74800799 | 0.15 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr5_-_173217931 | 0.13 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr17_+_74463650 | 0.12 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr3_-_141747950 | 0.12 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_169418195 | 0.12 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr7_-_150020578 | 0.11 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr20_-_33735070 | 0.11 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_-_25150373 | 0.11 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr1_+_225600404 | 0.11 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr6_+_114178512 | 0.10 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr17_+_1674982 | 0.10 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_-_124741451 | 0.10 |
ENST00000520519.1
|
ANXA13
|
annexin A13 |
| chr7_+_134528635 | 0.10 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr9_-_26947220 | 0.09 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr14_-_51027838 | 0.09 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_-_22862448 | 0.09 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr1_+_117544366 | 0.09 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr19_+_21324827 | 0.09 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
| chr16_-_66584059 | 0.09 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr2_+_152214098 | 0.09 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr7_+_5465382 | 0.08 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr19_-_51920952 | 0.08 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr12_+_26348246 | 0.08 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr16_-_28634874 | 0.08 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_-_36304201 | 0.08 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr4_+_158493642 | 0.08 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr12_+_4130143 | 0.08 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chr5_+_60933634 | 0.08 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr17_-_72772462 | 0.08 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_-_219850277 | 0.08 |
ENST00000295727.1
|
FEV
|
FEV (ETS oncogene family) |
| chr4_+_71019903 | 0.07 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr11_+_114168085 | 0.07 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr11_+_71934962 | 0.07 |
ENST00000543234.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr13_-_74993252 | 0.07 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr11_+_35222629 | 0.07 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_+_42989344 | 0.07 |
ENST00000244496.5
|
RRP36
|
ribosomal RNA processing 36 homolog (S. cerevisiae) |
| chr13_-_34250861 | 0.07 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr1_+_62439037 | 0.07 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr5_+_140762268 | 0.07 |
ENST00000518325.1
|
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr15_+_75970672 | 0.07 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr6_-_27835357 | 0.06 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr12_+_34175398 | 0.06 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chr14_+_103851712 | 0.06 |
ENST00000440884.3
ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3
|
MAP/microtubule affinity-regulating kinase 3 |
| chr9_-_215744 | 0.06 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr17_+_12569306 | 0.06 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr3_-_141719195 | 0.06 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_-_28937027 | 0.06 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr2_-_37068530 | 0.05 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr1_+_66820058 | 0.05 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr19_+_48949087 | 0.05 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr13_+_24144796 | 0.05 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr4_-_185275104 | 0.05 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr4_+_169418255 | 0.05 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr17_-_39093672 | 0.05 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr12_-_22063787 | 0.05 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr12_+_38710555 | 0.05 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr6_-_29527702 | 0.05 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr7_-_144435985 | 0.04 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr5_-_20575959 | 0.04 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr3_-_185538849 | 0.04 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_-_19887185 | 0.04 |
ENST00000590092.1
ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663
|
long intergenic non-protein coding RNA 663 |
| chr11_-_107729504 | 0.04 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_-_118796910 | 0.04 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr2_-_190927447 | 0.04 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr2_-_231860596 | 0.04 |
ENST00000441063.1
ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2
|
SPATA3 antisense RNA 1 (head to head) |
| chr10_-_25305011 | 0.04 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr17_+_12569472 | 0.04 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr11_-_128894053 | 0.04 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr4_-_177116772 | 0.04 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr15_+_69857515 | 0.04 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr2_-_241497374 | 0.04 |
ENST00000373318.2
ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chrX_+_47696337 | 0.04 |
ENST00000334937.4
ENST00000376950.4 |
ZNF81
|
zinc finger protein 81 |
| chr21_-_27423339 | 0.04 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr12_-_46121554 | 0.04 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr5_-_125930929 | 0.04 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr6_+_26087646 | 0.04 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr6_+_26087509 | 0.04 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr21_+_33671160 | 0.04 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_+_44840679 | 0.04 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr9_+_34652164 | 0.04 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr7_-_37024665 | 0.04 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_-_107729287 | 0.04 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr12_+_41136144 | 0.04 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr10_+_6779326 | 0.03 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr10_-_74283694 | 0.03 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr4_+_76481258 | 0.03 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr11_-_559377 | 0.03 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr19_+_21264980 | 0.03 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr12_-_10282836 | 0.03 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr2_-_74618964 | 0.03 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr5_+_95066823 | 0.03 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr2_-_241497390 | 0.03 |
ENST00000272972.3
ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr4_-_170948361 | 0.03 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_+_10509971 | 0.03 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr3_+_186692745 | 0.03 |
ENST00000438590.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_+_23945768 | 0.03 |
ENST00000486528.1
ENST00000496398.1 |
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr9_+_124329336 | 0.03 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr7_-_101272576 | 0.03 |
ENST00000223167.4
|
MYL10
|
myosin, light chain 10, regulatory |
| chr8_+_39972170 | 0.03 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr2_-_74619152 | 0.03 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr20_-_50418972 | 0.03 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr1_-_101360205 | 0.03 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr3_+_186353756 | 0.03 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr12_-_10978957 | 0.03 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr11_+_2405833 | 0.03 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr7_-_87856303 | 0.03 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr17_+_48823975 | 0.03 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr14_+_23025534 | 0.03 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr17_-_42992856 | 0.03 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr8_-_93978216 | 0.03 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chrX_-_100872911 | 0.03 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr12_+_49621658 | 0.03 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr12_-_118796971 | 0.03 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr1_-_153518270 | 0.03 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr3_+_39424828 | 0.03 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr20_-_50418947 | 0.02 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr6_-_136788001 | 0.02 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr5_-_41794313 | 0.02 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr10_+_18549645 | 0.02 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_+_18086392 | 0.02 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr9_-_77567743 | 0.02 |
ENST00000376854.5
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr21_-_22175341 | 0.02 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_-_92371839 | 0.02 |
ENST00000370399.2
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr17_+_57233087 | 0.02 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr8_-_57123815 | 0.02 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr20_+_30697298 | 0.02 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr16_+_28565230 | 0.02 |
ENST00000317058.3
|
CCDC101
|
coiled-coil domain containing 101 |
| chr12_+_59989918 | 0.02 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_+_212965170 | 0.02 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr7_+_30589829 | 0.02 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr19_-_3557570 | 0.02 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_+_75174204 | 0.02 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr6_+_26402465 | 0.02 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr6_+_55192267 | 0.02 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr3_-_114477787 | 0.02 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr3_-_114477962 | 0.02 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_+_129932974 | 0.02 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chrX_+_1710484 | 0.02 |
ENST00000313871.3
ENST00000381261.3 |
AKAP17A
|
A kinase (PRKA) anchor protein 17A |
| chr17_+_56833184 | 0.02 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr2_+_102615416 | 0.02 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr21_+_17909594 | 0.02 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_122854612 | 0.02 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr14_-_21994337 | 0.02 |
ENST00000537235.1
ENST00000450879.2 |
SALL2
|
spalt-like transcription factor 2 |
| chr10_-_101989315 | 0.02 |
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase |
| chr14_+_72052983 | 0.02 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_33686925 | 0.02 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_+_122610687 | 0.02 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr6_+_26365443 | 0.02 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr5_+_89770664 | 0.02 |
ENST00000503973.1
ENST00000399107.1 |
POLR3G
|
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr12_-_118797475 | 0.02 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr10_+_122610853 | 0.02 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr4_+_75174180 | 0.02 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr6_+_122793058 | 0.02 |
ENST00000392491.2
|
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr12_+_64798826 | 0.02 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr10_+_94451574 | 0.02 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr7_+_133812052 | 0.02 |
ENST00000285928.2
|
LRGUK
|
leucine-rich repeats and guanylate kinase domain containing |
| chr7_-_87342564 | 0.02 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr12_-_52779433 | 0.02 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr10_-_4285923 | 0.02 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr1_-_39347255 | 0.02 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr2_-_151395525 | 0.02 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr2_+_102456277 | 0.02 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr5_-_149829244 | 0.02 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr10_-_95241951 | 0.02 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_-_101039402 | 0.02 |
ENST00000193391.7
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr2_+_202047843 | 0.02 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr17_-_39203519 | 0.01 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr17_+_67498396 | 0.01 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_39033963 | 0.01 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chrX_-_13835147 | 0.01 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr8_-_52721975 | 0.01 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr4_+_87857538 | 0.01 |
ENST00000511442.1
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr7_-_87856280 | 0.01 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chrX_-_110655306 | 0.01 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr17_+_7155819 | 0.01 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr4_+_129731074 | 0.01 |
ENST00000512960.1
ENST00000503785.1 ENST00000514740.1 |
PHF17
|
jade family PHD finger 1 |
| chr6_-_10838710 | 0.01 |
ENST00000313243.2
|
MAK
|
male germ cell-associated kinase |
| chr6_-_133035185 | 0.01 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr12_+_26348429 | 0.01 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr9_+_71986182 | 0.01 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr6_-_10838736 | 0.01 |
ENST00000536370.1
ENST00000474039.1 |
MAK
|
male germ cell-associated kinase |
| chr9_+_125132803 | 0.01 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_-_176827631 | 0.01 |
ENST00000358571.2
|
PFN3
|
profilin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.0 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |