Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
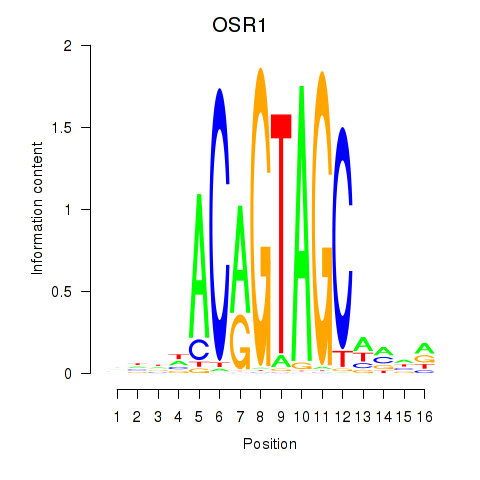
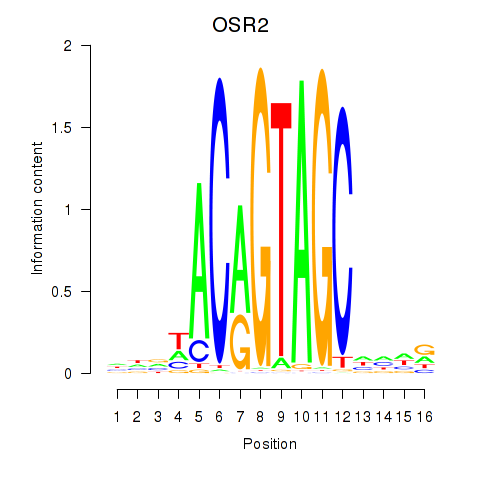
Results for OSR1_OSR2
Z-value: 0.76
Transcription factors associated with OSR1_OSR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OSR1
|
ENSG00000143867.5 | odd-skipped related transcription factor 1 |
|
OSR2
|
ENSG00000164920.5 | odd-skipped related transciption factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OSR2 | hg19_v2_chr8_+_99956662_99956686 | -0.49 | 5.1e-01 | Click! |
| OSR1 | hg19_v2_chr2_-_19558373_19558414 | 0.22 | 7.8e-01 | Click! |
Activity profile of OSR1_OSR2 motif
Sorted Z-values of OSR1_OSR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_185734773 | 0.62 |
ENST00000508020.1
|
RP11-701P16.2
|
Uncharacterized protein |
| chr15_+_63188009 | 0.58 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr15_-_74284558 | 0.40 |
ENST00000359750.4
ENST00000541638.1 ENST00000562453.1 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr6_-_15586238 | 0.39 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr18_+_52385068 | 0.38 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr1_-_153508460 | 0.34 |
ENST00000462776.2
|
S100A6
|
S100 calcium binding protein A6 |
| chr2_-_183387064 | 0.32 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_46690839 | 0.31 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr8_+_37594130 | 0.29 |
ENST00000518526.1
ENST00000523887.1 ENST00000276461.5 |
ERLIN2
|
ER lipid raft associated 2 |
| chr3_+_149192475 | 0.28 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_-_45457264 | 0.27 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chr19_+_19144384 | 0.25 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr1_-_151342180 | 0.25 |
ENST00000424475.1
|
SELENBP1
|
selenium binding protein 1 |
| chr3_+_10183447 | 0.24 |
ENST00000345392.2
|
VHL
|
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
| chr1_-_204463829 | 0.24 |
ENST00000429009.1
ENST00000415899.1 |
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr22_-_42336209 | 0.24 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr7_+_16700806 | 0.23 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr15_-_74284613 | 0.23 |
ENST00000316911.6
ENST00000564777.1 ENST00000566081.1 ENST00000316900.5 |
STOML1
|
stomatin (EPB72)-like 1 |
| chr7_+_134212312 | 0.23 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr22_+_39795746 | 0.23 |
ENST00000216160.6
ENST00000331454.3 |
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr13_-_52980263 | 0.23 |
ENST00000258613.4
ENST00000544466.1 |
THSD1
|
thrombospondin, type I, domain containing 1 |
| chr5_+_68463043 | 0.21 |
ENST00000508407.1
ENST00000505500.1 |
CCNB1
|
cyclin B1 |
| chr8_-_80942139 | 0.20 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr8_-_80942061 | 0.20 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr13_+_97874574 | 0.20 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr16_+_89696692 | 0.19 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr10_-_94301107 | 0.19 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr2_-_74007095 | 0.19 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr12_-_66035968 | 0.18 |
ENST00000537250.1
|
RP11-230G5.2
|
RP11-230G5.2 |
| chr14_+_56127960 | 0.18 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr12_-_50419177 | 0.18 |
ENST00000454520.2
ENST00000546595.1 ENST00000548824.1 ENST00000549777.1 ENST00000546723.1 ENST00000427314.2 ENST00000552157.1 ENST00000552310.1 ENST00000548644.1 ENST00000312377.5 ENST00000546786.1 ENST00000550149.1 ENST00000546764.1 ENST00000552004.1 ENST00000548320.1 ENST00000547905.1 ENST00000550651.1 ENST00000551145.1 ENST00000434422.1 ENST00000552921.1 |
RACGAP1
|
Rac GTPase activating protein 1 |
| chr17_-_72869086 | 0.17 |
ENST00000581530.1
ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR
|
ferredoxin reductase |
| chr4_+_169013666 | 0.17 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr1_+_12042015 | 0.17 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr8_+_66619277 | 0.17 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr2_+_87755054 | 0.16 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_13068012 | 0.15 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr5_-_125930929 | 0.15 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr6_+_10585979 | 0.15 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr22_-_23974506 | 0.15 |
ENST00000317749.5
|
C22orf43
|
chromosome 22 open reading frame 43 |
| chr1_-_156722195 | 0.15 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr8_+_96037255 | 0.14 |
ENST00000286687.4
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr7_+_76090993 | 0.14 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr19_-_41220957 | 0.14 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr17_+_60758814 | 0.14 |
ENST00000579432.1
ENST00000446119.2 |
MRC2
|
mannose receptor, C type 2 |
| chr19_+_17326521 | 0.13 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr1_+_144220127 | 0.13 |
ENST00000369373.5
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr17_-_72869140 | 0.13 |
ENST00000583917.1
ENST00000293195.5 ENST00000442102.2 |
FDXR
|
ferredoxin reductase |
| chr2_+_234959323 | 0.13 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr4_-_70080449 | 0.13 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr14_+_56127989 | 0.12 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr16_+_23690138 | 0.12 |
ENST00000300093.4
|
PLK1
|
polo-like kinase 1 |
| chr1_+_233749739 | 0.12 |
ENST00000366621.3
|
KCNK1
|
potassium channel, subfamily K, member 1 |
| chr5_-_125930877 | 0.12 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr15_+_79603404 | 0.12 |
ENST00000299705.5
|
TMED3
|
transmembrane emp24 protein transport domain containing 3 |
| chr4_+_146403912 | 0.12 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr2_+_234959376 | 0.11 |
ENST00000425558.1
|
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr6_-_136788001 | 0.11 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr22_-_42765174 | 0.11 |
ENST00000432473.1
ENST00000412060.1 ENST00000424852.1 |
Z83851.1
|
Z83851.1 |
| chr5_+_169011033 | 0.11 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr14_-_93651186 | 0.11 |
ENST00000556883.1
ENST00000298894.4 |
MOAP1
|
modulator of apoptosis 1 |
| chr22_-_45559540 | 0.11 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr12_+_94551874 | 0.11 |
ENST00000551850.1
|
PLXNC1
|
plexin C1 |
| chr3_+_149191723 | 0.11 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr19_-_16770915 | 0.11 |
ENST00000593459.1
ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4
SMIM7
|
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr14_-_107083690 | 0.10 |
ENST00000455737.1
ENST00000390629.2 |
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr10_-_81742364 | 0.10 |
ENST00000444384.3
|
SFTPD
|
surfactant protein D |
| chr5_+_68462837 | 0.10 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr5_+_110073853 | 0.10 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr1_-_231560790 | 0.10 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr4_-_77996032 | 0.10 |
ENST00000505609.1
|
CCNI
|
cyclin I |
| chr19_-_56709162 | 0.10 |
ENST00000589938.1
ENST00000587032.2 ENST00000586855.2 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr8_+_125486939 | 0.10 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr14_+_105267250 | 0.10 |
ENST00000342537.7
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr17_-_79895097 | 0.09 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr8_+_26247878 | 0.09 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr7_-_6388537 | 0.09 |
ENST00000313324.4
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220, member A |
| chr11_-_63439381 | 0.09 |
ENST00000538786.1
ENST00000540699.1 |
ATL3
|
atlastin GTPase 3 |
| chr19_+_19144666 | 0.09 |
ENST00000535288.1
ENST00000538663.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr20_-_30311703 | 0.09 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr22_+_42017280 | 0.09 |
ENST00000402580.3
ENST00000428575.2 ENST00000359308.4 |
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr22_+_41763274 | 0.09 |
ENST00000406644.3
|
TEF
|
thyrotrophic embryonic factor |
| chr19_+_7694623 | 0.09 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr3_+_50654821 | 0.09 |
ENST00000457064.1
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr13_+_53226963 | 0.09 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr12_-_123201337 | 0.09 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr21_+_40752170 | 0.09 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr3_-_50360192 | 0.09 |
ENST00000442581.1
ENST00000447092.1 ENST00000357750.4 |
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr6_+_80341000 | 0.08 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr1_+_26503894 | 0.08 |
ENST00000361530.6
ENST00000374253.5 |
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr20_-_33999766 | 0.08 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr11_-_63439013 | 0.08 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr16_-_66968055 | 0.08 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr16_+_88493879 | 0.08 |
ENST00000565624.1
ENST00000437464.1 |
ZNF469
|
zinc finger protein 469 |
| chr16_+_67143880 | 0.08 |
ENST00000219139.3
ENST00000566026.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr11_+_64808675 | 0.08 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr11_+_8704748 | 0.08 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr12_-_58165870 | 0.08 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr11_+_62648336 | 0.07 |
ENST00000338663.7
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr5_+_68462944 | 0.07 |
ENST00000506572.1
|
CCNB1
|
cyclin B1 |
| chr22_+_42017459 | 0.07 |
ENST00000405878.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr16_-_8962200 | 0.07 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr1_+_161736072 | 0.07 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_-_32860020 | 0.07 |
ENST00000527163.1
ENST00000341071.7 ENST00000530485.1 ENST00000446293.2 ENST00000413080.1 ENST00000449308.1 ENST00000526031.1 ENST00000419121.2 ENST00000455895.2 |
BSDC1
|
BSD domain containing 1 |
| chr21_-_34915084 | 0.07 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr19_+_36203830 | 0.07 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr12_+_94656297 | 0.07 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr10_+_97515409 | 0.07 |
ENST00000371207.3
ENST00000543964.1 |
ENTPD1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr2_+_70056762 | 0.07 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr1_+_196743943 | 0.07 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr11_-_63439174 | 0.06 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr1_+_89829610 | 0.06 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr10_+_78078088 | 0.06 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr15_+_75491213 | 0.06 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr12_+_27396901 | 0.06 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr12_-_130529501 | 0.06 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr19_-_17185848 | 0.06 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr19_+_30719410 | 0.06 |
ENST00000585628.1
ENST00000591488.1 |
ZNF536
|
zinc finger protein 536 |
| chr17_-_79895154 | 0.06 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_-_41694640 | 0.06 |
ENST00000558719.1
ENST00000260361.4 ENST00000560978.1 |
NDUFAF1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr11_-_35287243 | 0.05 |
ENST00000464522.2
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_-_185733394 | 0.05 |
ENST00000504342.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr10_+_99400443 | 0.05 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr14_-_39417410 | 0.05 |
ENST00000557019.1
ENST00000556116.1 ENST00000554732.1 |
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr14_+_21152706 | 0.05 |
ENST00000397995.2
ENST00000304704.4 ENST00000553909.1 |
RNASE4
AL163636.6
|
ribonuclease, RNase A family, 4 Homo sapiens ribonuclease, RNase A family, 4 (RNASE4), transcript variant 4, mRNA. |
| chr1_+_156561533 | 0.05 |
ENST00000368234.3
ENST00000368235.3 ENST00000368233.3 |
APOA1BP
|
apolipoprotein A-I binding protein |
| chr2_+_217277466 | 0.05 |
ENST00000358207.5
ENST00000434435.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr1_+_201798269 | 0.05 |
ENST00000361565.4
|
IPO9
|
importin 9 |
| chr17_+_40714092 | 0.04 |
ENST00000420359.1
ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY
|
CoA synthase |
| chr5_+_169010638 | 0.04 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr1_+_10003486 | 0.04 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr20_+_31407692 | 0.04 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr2_+_178257372 | 0.04 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr14_-_39417473 | 0.04 |
ENST00000553932.1
|
LINC00639
|
long intergenic non-protein coding RNA 639 |
| chr7_+_127228399 | 0.04 |
ENST00000000233.5
ENST00000415666.1 |
ARF5
|
ADP-ribosylation factor 5 |
| chr4_-_122791583 | 0.04 |
ENST00000506636.1
ENST00000264499.4 |
BBS7
|
Bardet-Biedl syndrome 7 |
| chr9_-_95055956 | 0.04 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr15_+_41523417 | 0.03 |
ENST00000560397.1
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr17_-_191188 | 0.03 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr9_-_98268883 | 0.03 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr4_+_69681710 | 0.03 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr7_-_105752971 | 0.03 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chrX_+_9880590 | 0.03 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr8_+_94710789 | 0.03 |
ENST00000523475.1
|
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr14_+_101359265 | 0.03 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr2_-_74735707 | 0.03 |
ENST00000233630.6
|
PCGF1
|
polycomb group ring finger 1 |
| chr3_-_186080012 | 0.03 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr6_+_32938665 | 0.03 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr8_-_125486755 | 0.03 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chrX_+_80457442 | 0.03 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr6_+_3982909 | 0.03 |
ENST00000356722.3
|
C6ORF50
|
C6ORF50 |
| chr14_+_21152259 | 0.03 |
ENST00000555835.1
ENST00000336811.6 |
RNASE4
ANG
|
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chrX_-_154060946 | 0.03 |
ENST00000369529.1
|
SMIM9
|
small integral membrane protein 9 |
| chr1_+_196857144 | 0.03 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr17_-_79894651 | 0.03 |
ENST00000584848.1
ENST00000577756.1 ENST00000329875.8 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_-_33864772 | 0.02 |
ENST00000361112.4
|
SLFN12L
|
schlafen family member 12-like |
| chr18_+_63418068 | 0.02 |
ENST00000397968.2
|
CDH7
|
cadherin 7, type 2 |
| chr21_+_43919710 | 0.02 |
ENST00000398341.3
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr12_-_14967095 | 0.02 |
ENST00000316048.2
|
SMCO3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr10_-_53459319 | 0.02 |
ENST00000331173.4
|
CSTF2T
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr17_+_61271355 | 0.02 |
ENST00000583356.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_-_88125471 | 0.02 |
ENST00000398146.3
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr19_-_52391142 | 0.02 |
ENST00000446514.1
ENST00000458390.1 ENST00000420592.1 ENST00000451628.2 ENST00000592321.1 ENST00000591320.1 ENST00000412216.1 ENST00000301399.5 |
ZNF577
|
zinc finger protein 577 |
| chr11_-_77705687 | 0.02 |
ENST00000529807.1
ENST00000527522.1 ENST00000534064.1 |
INTS4
|
integrator complex subunit 4 |
| chr2_-_183387283 | 0.02 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr9_-_95056010 | 0.02 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr4_+_145567173 | 0.02 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr1_-_25291475 | 0.02 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr2_-_44588679 | 0.02 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr1_-_207143802 | 0.02 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr2_-_44588694 | 0.02 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr9_+_36572851 | 0.02 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr8_-_99306611 | 0.02 |
ENST00000341166.3
|
NIPAL2
|
NIPA-like domain containing 2 |
| chr15_-_66649010 | 0.02 |
ENST00000367709.4
ENST00000261881.4 |
TIPIN
|
TIMELESS interacting protein |
| chr11_+_8704298 | 0.02 |
ENST00000531978.1
ENST00000524496.1 ENST00000532359.1 ENST00000530022.1 |
RPL27A
|
ribosomal protein L27a |
| chr3_+_130648842 | 0.02 |
ENST00000508297.1
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr10_-_43904608 | 0.02 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_+_196743912 | 0.02 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr20_-_1472029 | 0.02 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr21_-_34915147 | 0.01 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr3_+_40498783 | 0.01 |
ENST00000338970.6
ENST00000396203.2 ENST00000416518.1 |
RPL14
|
ribosomal protein L14 |
| chr4_+_6576895 | 0.01 |
ENST00000285599.3
ENST00000504248.1 ENST00000505907.1 |
MAN2B2
|
mannosidase, alpha, class 2B, member 2 |
| chr16_-_2827128 | 0.01 |
ENST00000494946.2
ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr11_+_65407331 | 0.01 |
ENST00000527525.1
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr8_+_104384616 | 0.01 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr10_-_43904235 | 0.01 |
ENST00000356053.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr7_-_144533074 | 0.01 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr22_+_24115000 | 0.01 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr3_+_156799587 | 0.01 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr3_+_101504200 | 0.01 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr22_+_32870661 | 0.01 |
ENST00000266087.7
|
FBXO7
|
F-box protein 7 |
| chr15_-_60683326 | 0.01 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr14_+_65453432 | 0.01 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr14_+_22458631 | 0.01 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr10_+_95848824 | 0.01 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr19_+_19303008 | 0.01 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr11_-_82997420 | 0.01 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chrX_-_134049233 | 0.01 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr5_-_124081008 | 0.01 |
ENST00000306315.5
|
ZNF608
|
zinc finger protein 608 |
| chr6_+_122720681 | 0.01 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OSR1_OSR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000775 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.1 | 0.2 | GO:0016488 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.1 | 0.3 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004324 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |