Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
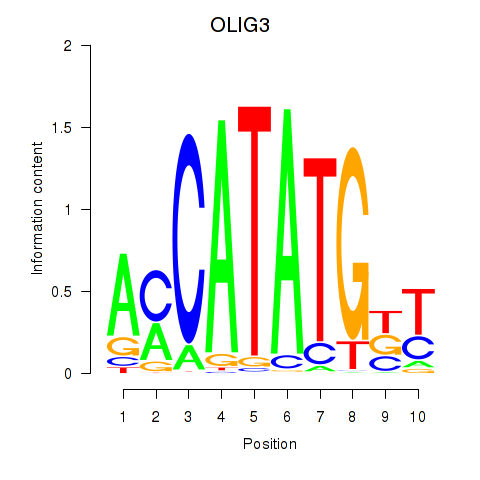
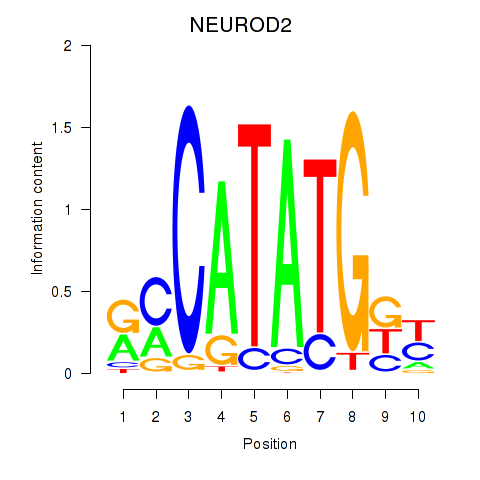
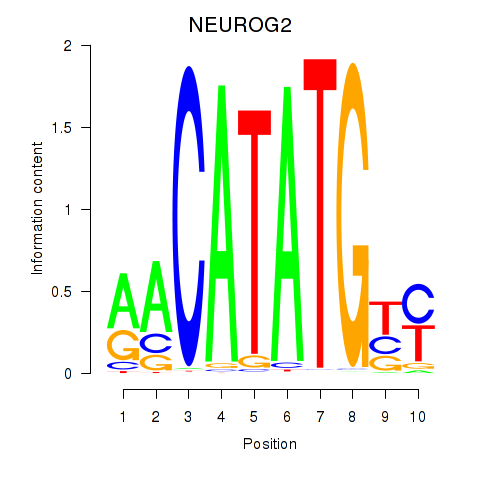
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 0.72
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.5 | oligodendrocyte transcription factor 3 |
|
NEUROD2
|
ENSG00000171532.4 | neuronal differentiation 2 |
|
NEUROG2
|
ENSG00000178403.3 | neurogenin 2 |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_142264664 | 0.85 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr2_+_66918558 | 0.58 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr12_+_2912363 | 0.56 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr17_-_47925379 | 0.56 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr3_+_72201910 | 0.52 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr2_-_132589601 | 0.52 |
ENST00000437330.1
|
AC103564.7
|
AC103564.7 |
| chr14_-_69444957 | 0.43 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr2_+_90458201 | 0.39 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr13_+_49551020 | 0.37 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr3_+_112930387 | 0.34 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr12_+_42624050 | 0.33 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chr6_+_30539153 | 0.32 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr9_-_130635741 | 0.32 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr5_-_111312622 | 0.32 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr6_+_84563295 | 0.31 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr6_+_31553978 | 0.31 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr3_+_137717571 | 0.29 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr3_-_186524144 | 0.29 |
ENST00000427785.1
|
RFC4
|
replication factor C (activator 1) 4, 37kDa |
| chr11_-_115127611 | 0.28 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr10_+_51565108 | 0.27 |
ENST00000438493.1
ENST00000452682.1 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr10_+_51565188 | 0.27 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr8_+_39972170 | 0.27 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr4_-_120243545 | 0.27 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr4_-_129491686 | 0.27 |
ENST00000514265.1
|
RP11-184M15.1
|
RP11-184M15.1 |
| chr2_-_158345462 | 0.26 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr6_-_160147925 | 0.25 |
ENST00000535561.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr18_+_44812072 | 0.25 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr4_+_54927213 | 0.25 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr3_-_129375556 | 0.25 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr10_-_104597286 | 0.24 |
ENST00000369887.3
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr11_-_62358972 | 0.24 |
ENST00000278279.3
|
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr5_+_71475449 | 0.23 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr19_+_42041860 | 0.23 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr8_-_95449155 | 0.23 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr21_-_43786634 | 0.22 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr8_-_135522425 | 0.21 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr7_-_97881429 | 0.21 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr10_+_99627889 | 0.20 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr7_+_105172612 | 0.20 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr2_-_113999260 | 0.20 |
ENST00000468980.2
|
PAX8
|
paired box 8 |
| chr16_+_68279256 | 0.20 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr2_-_154335300 | 0.20 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr7_-_41742697 | 0.20 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr7_-_24957699 | 0.19 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr5_+_175299743 | 0.19 |
ENST00000502265.1
|
CPLX2
|
complexin 2 |
| chr21_+_42792442 | 0.19 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr18_-_2982869 | 0.19 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr15_+_74466012 | 0.19 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr6_+_24126350 | 0.18 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr20_-_36794938 | 0.17 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr16_+_67280799 | 0.17 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr7_+_134430212 | 0.17 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr15_+_40650408 | 0.17 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr1_+_95975672 | 0.17 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr1_+_225600404 | 0.17 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr6_+_13182751 | 0.17 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr12_-_113658826 | 0.16 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr4_+_160188889 | 0.16 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr18_-_47814032 | 0.16 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr6_+_155585147 | 0.16 |
ENST00000367165.3
|
CLDN20
|
claudin 20 |
| chr1_+_15479054 | 0.15 |
ENST00000376014.3
ENST00000451326.2 |
TMEM51
|
transmembrane protein 51 |
| chr3_-_53164423 | 0.15 |
ENST00000467048.1
ENST00000394738.3 ENST00000296292.3 |
RFT1
|
RFT1 homolog (S. cerevisiae) |
| chr2_-_158295915 | 0.15 |
ENST00000418920.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr10_+_133918175 | 0.15 |
ENST00000298622.4
|
JAKMIP3
|
Janus kinase and microtubule interacting protein 3 |
| chr15_+_75940218 | 0.14 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr9_-_35361262 | 0.14 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chrX_-_13835461 | 0.14 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr5_-_171433579 | 0.14 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr10_+_44101850 | 0.14 |
ENST00000361807.3
ENST00000374437.2 ENST00000430885.1 ENST00000374435.3 |
ZNF485
|
zinc finger protein 485 |
| chr4_-_7873713 | 0.14 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr12_+_41136144 | 0.14 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr5_+_135383008 | 0.14 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr6_+_37400974 | 0.14 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr2_-_97760576 | 0.14 |
ENST00000414820.1
ENST00000272610.3 |
FAHD2B
|
fumarylacetoacetate hydrolase domain containing 2B |
| chr11_+_134123389 | 0.14 |
ENST00000281182.4
ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8
|
acyl-CoA dehydrogenase family, member 8 |
| chrX_+_118108601 | 0.14 |
ENST00000371628.3
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr11_-_47616210 | 0.14 |
ENST00000302514.3
|
C1QTNF4
|
C1q and tumor necrosis factor related protein 4 |
| chr15_+_63354769 | 0.14 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr9_+_5450503 | 0.13 |
ENST00000381573.4
ENST00000381577.3 |
CD274
|
CD274 molecule |
| chr3_-_27410847 | 0.13 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr1_-_168464875 | 0.13 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr11_-_11374904 | 0.13 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chrX_+_118108571 | 0.13 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr18_-_6928495 | 0.13 |
ENST00000580197.1
|
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr1_+_206808868 | 0.13 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr2_+_64068116 | 0.13 |
ENST00000480679.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr3_+_38307313 | 0.13 |
ENST00000450935.2
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr2_-_106777040 | 0.13 |
ENST00000444193.1
ENST00000416298.1 |
UXS1
|
UDP-glucuronate decarboxylase 1 |
| chr17_+_77681075 | 0.13 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr1_+_206808918 | 0.13 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr16_-_47493041 | 0.13 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr20_-_36794902 | 0.13 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr11_+_65405556 | 0.12 |
ENST00000534313.1
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr16_-_21877629 | 0.12 |
ENST00000544693.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr8_-_41508855 | 0.12 |
ENST00000518699.2
|
NKX6-3
|
NK6 homeobox 3 |
| chr5_-_137674000 | 0.12 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr1_+_6511651 | 0.12 |
ENST00000434576.1
|
ESPN
|
espin |
| chr6_-_28891709 | 0.12 |
ENST00000377194.3
ENST00000377199.3 |
TRIM27
|
tripartite motif containing 27 |
| chr12_-_75723805 | 0.12 |
ENST00000409799.1
ENST00000409445.3 |
CAPS2
|
calcyphosine 2 |
| chr22_+_22723969 | 0.11 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr19_-_44171817 | 0.11 |
ENST00000593714.1
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr11_+_32112431 | 0.11 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr3_+_98699880 | 0.11 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr4_-_174451370 | 0.11 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr3_+_113251143 | 0.11 |
ENST00000264852.4
ENST00000393830.3 |
SIDT1
|
SID1 transmembrane family, member 1 |
| chr22_-_30953587 | 0.11 |
ENST00000453479.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr15_+_67418047 | 0.11 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr3_+_185300391 | 0.11 |
ENST00000545472.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr18_+_61144160 | 0.11 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr19_+_44764031 | 0.11 |
ENST00000592581.1
ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233
|
zinc finger protein 233 |
| chr12_+_113416191 | 0.11 |
ENST00000342315.4
ENST00000392583.2 |
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr15_+_41099254 | 0.10 |
ENST00000570108.1
ENST00000564258.1 ENST00000355341.4 ENST00000336455.5 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr1_+_244214577 | 0.10 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr4_+_77941685 | 0.10 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr19_-_36705547 | 0.10 |
ENST00000304116.5
|
ZNF565
|
zinc finger protein 565 |
| chr4_-_24914576 | 0.10 |
ENST00000502801.1
ENST00000428116.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr1_-_169396666 | 0.10 |
ENST00000456107.1
ENST00000367805.3 |
CCDC181
|
coiled-coil domain containing 181 |
| chr1_-_156722195 | 0.10 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_-_158345341 | 0.10 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr10_+_88728189 | 0.10 |
ENST00000416348.1
|
ADIRF
|
adipogenesis regulatory factor |
| chr5_-_150466692 | 0.10 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr7_-_112758665 | 0.10 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_+_116502605 | 0.10 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr1_+_201592013 | 0.10 |
ENST00000593583.1
|
AC096677.1
|
Uncharacterized protein ENSP00000471857 |
| chr10_-_27389392 | 0.10 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr5_-_150460539 | 0.10 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr12_+_113376249 | 0.10 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr2_+_191792376 | 0.10 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr1_-_40098672 | 0.10 |
ENST00000535435.1
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr18_+_21572737 | 0.10 |
ENST00000304621.6
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr4_-_40632140 | 0.10 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr2_-_214148921 | 0.10 |
ENST00000360083.3
|
AC079610.2
|
AC079610.2 |
| chr14_-_75078725 | 0.09 |
ENST00000556690.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr1_-_45272951 | 0.09 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr3_-_178984759 | 0.09 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr11_-_110167352 | 0.09 |
ENST00000533991.1
ENST00000528498.1 ENST00000405097.1 ENST00000528900.1 ENST00000530301.1 ENST00000343115.4 |
RDX
|
radixin |
| chr15_+_71389281 | 0.09 |
ENST00000355327.3
|
THSD4
|
thrombospondin, type I, domain containing 4 |
| chr17_+_46125685 | 0.09 |
ENST00000579889.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr4_+_174818390 | 0.09 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr17_+_80858418 | 0.09 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr11_-_47870091 | 0.09 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr11_-_115158193 | 0.09 |
ENST00000543540.1
|
CADM1
|
cell adhesion molecule 1 |
| chr3_+_127323739 | 0.09 |
ENST00000491422.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr6_-_150219232 | 0.09 |
ENST00000531073.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr14_-_25479811 | 0.09 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr17_+_46048497 | 0.09 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr18_+_3252206 | 0.09 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_56862301 | 0.09 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr8_+_103540983 | 0.09 |
ENST00000523572.1
|
KB-1980E6.3
|
Uncharacterized protein |
| chr11_+_70049269 | 0.09 |
ENST00000301838.4
|
FADD
|
Fas (TNFRSF6)-associated via death domain |
| chr6_-_150212029 | 0.09 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr16_-_67965756 | 0.08 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr3_-_25915189 | 0.08 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr17_+_56769924 | 0.08 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chrX_-_77225135 | 0.08 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr7_-_45957011 | 0.08 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr18_+_3252265 | 0.08 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_10331605 | 0.08 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chrX_-_53310791 | 0.08 |
ENST00000375365.2
|
IQSEC2
|
IQ motif and Sec7 domain 2 |
| chr3_+_193809263 | 0.08 |
ENST00000457815.1
|
RP11-135A1.3
|
RP11-135A1.3 |
| chr6_+_150690133 | 0.08 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr19_-_37019562 | 0.08 |
ENST00000523638.1
|
ZNF260
|
zinc finger protein 260 |
| chr3_+_141043050 | 0.08 |
ENST00000509842.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_+_113667354 | 0.08 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr12_+_52643077 | 0.08 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr4_+_109571740 | 0.08 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr17_-_73874654 | 0.08 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr2_+_201987200 | 0.08 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_+_58166726 | 0.08 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr4_-_22444733 | 0.08 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr17_-_40170506 | 0.08 |
ENST00000589773.1
ENST00000590348.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr17_-_19281203 | 0.08 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr3_+_63638372 | 0.08 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr6_+_3000057 | 0.08 |
ENST00000397717.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr5_-_150467221 | 0.08 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr2_+_201994042 | 0.08 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_177053307 | 0.08 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr6_+_36973406 | 0.08 |
ENST00000274963.8
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr19_-_10213335 | 0.08 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr3_-_171489085 | 0.08 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr2_+_201242715 | 0.08 |
ENST00000421573.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_15479021 | 0.08 |
ENST00000428417.1
|
TMEM51
|
transmembrane protein 51 |
| chr22_-_36556821 | 0.08 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr12_-_2944184 | 0.08 |
ENST00000337508.4
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr19_+_16999654 | 0.08 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr9_-_111775772 | 0.07 |
ENST00000325580.6
ENST00000374593.4 ENST00000374595.4 ENST00000325551.4 |
CTNNAL1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr6_+_151561506 | 0.07 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_-_165424973 | 0.07 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr1_-_232598163 | 0.07 |
ENST00000308942.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_+_151561085 | 0.07 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chrX_-_46187069 | 0.07 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr1_-_226076843 | 0.07 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr11_-_125648690 | 0.07 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr2_+_85661918 | 0.07 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr15_+_62853562 | 0.07 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr3_+_48507621 | 0.07 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr19_-_21512144 | 0.07 |
ENST00000602023.1
|
ZNF708
|
zinc finger protein 708 |
| chr3_-_43147549 | 0.07 |
ENST00000344697.2
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr5_-_36242119 | 0.07 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr2_+_219110149 | 0.07 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.2 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.2 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.3 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0033319 | UDP-D-xylose metabolic process(GO:0033319) UDP-D-xylose biosynthetic process(GO:0033320) |
| 0.0 | 0.3 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.1 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0061032 | cardiac right ventricle formation(GO:0003219) noradrenergic neuron differentiation(GO:0003357) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.0 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.0 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.3 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) regulation of response to drug(GO:2001023) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.0 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0048040 | UDP-glucuronate decarboxylase activity(GO:0048040) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |