Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
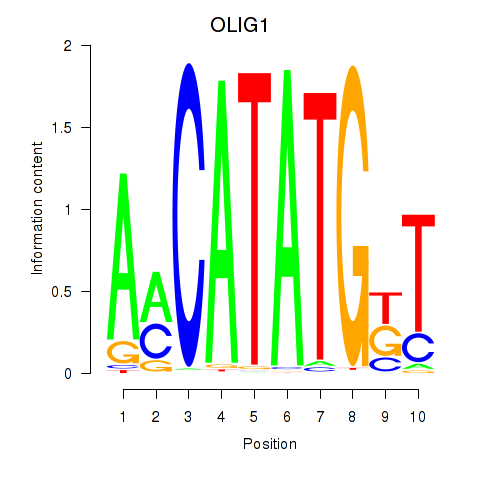
Results for OLIG1
Z-value: 0.53
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.8 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OLIG1 | hg19_v2_chr21_+_34442439_34442455 | -0.46 | 5.4e-01 | Click! |
Activity profile of OLIG1 motif
Sorted Z-values of OLIG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_32414059 | 0.67 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr1_+_76251912 | 0.57 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_+_90458201 | 0.56 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr4_+_54927213 | 0.55 |
ENST00000595906.1
|
AC110792.1
|
HCG2027126; Uncharacterized protein |
| chr3_-_148939598 | 0.49 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_129375556 | 0.46 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr19_+_42041860 | 0.43 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr19_-_58204128 | 0.43 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr19_-_39368887 | 0.35 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr5_+_68485433 | 0.35 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr4_+_77941685 | 0.34 |
ENST00000506731.1
|
SEPT11
|
septin 11 |
| chr13_-_46679185 | 0.30 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr17_-_39156138 | 0.29 |
ENST00000391587.1
|
KRTAP3-2
|
keratin associated protein 3-2 |
| chr12_+_133657461 | 0.28 |
ENST00000412146.2
ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140
|
zinc finger protein 140 |
| chr5_-_114632307 | 0.28 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr1_+_186344883 | 0.28 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr6_-_52859046 | 0.27 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_-_114631958 | 0.27 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr1_+_186344945 | 0.25 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr12_-_8088773 | 0.24 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr7_+_116502605 | 0.24 |
ENST00000458284.2
ENST00000490693.1 |
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr4_-_159094194 | 0.24 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr1_-_45253377 | 0.24 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr3_-_46068969 | 0.23 |
ENST00000542109.1
ENST00000395946.2 |
XCR1
|
chemokine (C motif) receptor 1 |
| chr5_+_68485363 | 0.23 |
ENST00000283006.2
ENST00000515001.1 |
CENPH
|
centromere protein H |
| chr3_+_72201910 | 0.22 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr15_-_38519066 | 0.22 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr8_+_101349823 | 0.21 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr14_-_55658323 | 0.21 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr4_+_109571740 | 0.21 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr16_+_12059091 | 0.20 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_-_148939835 | 0.20 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_-_71842941 | 0.20 |
ENST00000423132.2
ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr3_+_113667354 | 0.20 |
ENST00000491556.1
|
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr17_+_39846114 | 0.20 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chrX_-_84363974 | 0.19 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr15_-_55700216 | 0.19 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr18_-_59274139 | 0.18 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr3_-_121379739 | 0.18 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_91573132 | 0.17 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr8_+_62747349 | 0.17 |
ENST00000517953.1
ENST00000520097.1 ENST00000519766.1 |
RP11-705O24.1
|
RP11-705O24.1 |
| chr10_+_89124746 | 0.17 |
ENST00000465545.1
|
NUTM2D
|
NUT family member 2D |
| chr11_+_120107344 | 0.17 |
ENST00000260264.4
|
POU2F3
|
POU class 2 homeobox 3 |
| chr7_-_112758665 | 0.17 |
ENST00000397764.3
|
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr7_-_92146729 | 0.17 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr20_+_47662805 | 0.17 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr3_-_195310802 | 0.16 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chrX_+_69509870 | 0.16 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr9_-_119162885 | 0.16 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr9_-_35361262 | 0.16 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chr4_-_141348763 | 0.16 |
ENST00000509477.1
|
CLGN
|
calmegin |
| chr1_+_153750622 | 0.16 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr7_+_107531580 | 0.16 |
ENST00000537148.1
ENST00000440410.1 ENST00000437604.2 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr14_-_107283278 | 0.16 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr10_+_51565108 | 0.16 |
ENST00000438493.1
ENST00000452682.1 |
NCOA4
|
nuclear receptor coactivator 4 |
| chrX_-_11129229 | 0.15 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr10_+_51565188 | 0.15 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr14_-_55658252 | 0.14 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr16_-_71843047 | 0.14 |
ENST00000299980.4
ENST00000393512.3 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr14_+_38091270 | 0.14 |
ENST00000553443.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr22_-_19435755 | 0.14 |
ENST00000542103.1
ENST00000399562.4 |
C22orf39
|
chromosome 22 open reading frame 39 |
| chr3_-_98241358 | 0.14 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr4_+_146402843 | 0.14 |
ENST00000514831.1
|
SMAD1
|
SMAD family member 1 |
| chr2_+_75873902 | 0.14 |
ENST00000393909.2
ENST00000358788.6 ENST00000409374.1 |
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr2_-_228582709 | 0.14 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr1_+_91966656 | 0.14 |
ENST00000428239.1
ENST00000426137.1 |
CDC7
|
cell division cycle 7 |
| chr8_+_48173556 | 0.14 |
ENST00000524006.1
|
SPIDR
|
scaffolding protein involved in DNA repair |
| chr4_+_71600063 | 0.14 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr5_+_135496675 | 0.13 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr1_+_203274639 | 0.13 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr10_-_75168071 | 0.13 |
ENST00000394847.3
|
ANXA7
|
annexin A7 |
| chr17_+_42923686 | 0.13 |
ENST00000591513.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr10_+_180405 | 0.13 |
ENST00000439456.1
ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr2_-_182545603 | 0.13 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr5_-_75919217 | 0.13 |
ENST00000504899.1
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr3_+_150321068 | 0.13 |
ENST00000471696.1
ENST00000477889.1 ENST00000485923.1 |
SELT
|
Selenoprotein T |
| chr17_-_47925379 | 0.12 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr17_+_44790515 | 0.12 |
ENST00000576346.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr2_-_188378368 | 0.12 |
ENST00000392365.1
ENST00000435414.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_+_205538105 | 0.12 |
ENST00000367147.4
ENST00000539267.1 |
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr1_+_206808868 | 0.12 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr3_+_112709804 | 0.12 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr13_+_28813645 | 0.12 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr16_+_68279256 | 0.12 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr2_+_183989157 | 0.12 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr10_-_69597828 | 0.12 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr4_-_74088800 | 0.11 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr12_-_94673956 | 0.11 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr5_-_75919253 | 0.11 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr1_+_220863187 | 0.11 |
ENST00000294889.5
|
C1orf115
|
chromosome 1 open reading frame 115 |
| chr8_+_35649365 | 0.11 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr10_-_11574274 | 0.11 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chrX_+_120181457 | 0.11 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr4_-_89744457 | 0.11 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_-_63448973 | 0.11 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr3_+_62304648 | 0.11 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr7_-_11871815 | 0.11 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr10_-_69597915 | 0.10 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr11_+_63449045 | 0.10 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr19_-_11450249 | 0.10 |
ENST00000222120.3
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr10_-_15902449 | 0.10 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr19_+_12035913 | 0.10 |
ENST00000591944.1
|
ZNF763
|
Uncharacterized protein; Zinc finger protein 763 |
| chr3_+_113666748 | 0.10 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr4_-_153456153 | 0.10 |
ENST00000603548.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_169764163 | 0.09 |
ENST00000413811.2
ENST00000359326.4 ENST00000456684.1 |
C1orf112
|
chromosome 1 open reading frame 112 |
| chr2_+_75874105 | 0.09 |
ENST00000476622.1
|
MRPL19
|
mitochondrial ribosomal protein L19 |
| chr2_-_32490801 | 0.09 |
ENST00000360906.5
ENST00000342905.6 |
NLRC4
|
NLR family, CARD domain containing 4 |
| chr16_-_69385681 | 0.09 |
ENST00000288025.3
|
TMED6
|
transmembrane emp24 protein transport domain containing 6 |
| chr1_+_87458692 | 0.09 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr10_-_69597810 | 0.09 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chrX_+_101975619 | 0.09 |
ENST00000457056.1
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr7_-_112758589 | 0.09 |
ENST00000413744.1
ENST00000439551.1 ENST00000441359.1 |
LINC00998
|
long intergenic non-protein coding RNA 998 |
| chr1_+_206808918 | 0.09 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr11_-_44972418 | 0.09 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr10_-_5446786 | 0.09 |
ENST00000479328.1
ENST00000380419.3 |
TUBAL3
|
tubulin, alpha-like 3 |
| chrX_+_101975643 | 0.09 |
ENST00000361229.4
|
BHLHB9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr8_+_21823726 | 0.08 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr8_-_38239732 | 0.08 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr1_+_110881945 | 0.08 |
ENST00000602849.1
ENST00000487146.2 |
RBM15
|
RNA binding motif protein 15 |
| chr15_-_45670924 | 0.08 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr19_+_40873617 | 0.08 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr10_-_94257512 | 0.08 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr1_+_81106951 | 0.08 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr8_-_95449155 | 0.08 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_-_101491319 | 0.08 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr6_+_57182400 | 0.08 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr17_+_80858418 | 0.08 |
ENST00000574422.1
|
TBCD
|
tubulin folding cofactor D |
| chr6_-_25874440 | 0.08 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr8_-_142012169 | 0.08 |
ENST00000517453.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr10_-_1071796 | 0.08 |
ENST00000277517.1
|
IDI2
|
isopentenyl-diphosphate delta isomerase 2 |
| chr2_+_191792376 | 0.08 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr8_+_95835438 | 0.08 |
ENST00000521860.1
ENST00000519457.1 ENST00000519053.1 ENST00000523731.1 ENST00000447247.1 |
INTS8
|
integrator complex subunit 8 |
| chr8_-_102181718 | 0.08 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr1_+_173837214 | 0.08 |
ENST00000367704.1
|
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr6_+_4087664 | 0.08 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr4_+_75858318 | 0.08 |
ENST00000307428.7
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chrX_+_107334983 | 0.07 |
ENST00000457035.1
ENST00000545696.1 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr7_-_24957699 | 0.07 |
ENST00000441059.1
ENST00000415162.1 |
OSBPL3
|
oxysterol binding protein-like 3 |
| chr15_-_49103235 | 0.07 |
ENST00000380950.2
|
CEP152
|
centrosomal protein 152kDa |
| chr5_-_133510456 | 0.07 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr4_-_89744314 | 0.07 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chrX_-_102757802 | 0.07 |
ENST00000372633.1
|
RAB40A
|
RAB40A, member RAS oncogene family |
| chr14_+_19377522 | 0.07 |
ENST00000550708.1
|
OR11H12
|
olfactory receptor, family 11, subfamily H, member 12 |
| chr4_-_89744365 | 0.07 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_+_59726565 | 0.07 |
ENST00000412930.2
|
FKSG52
|
FKSG52 |
| chr8_-_23315190 | 0.07 |
ENST00000356206.6
ENST00000358689.4 ENST00000417069.2 |
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr10_+_180987 | 0.06 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr15_+_65843130 | 0.06 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_-_197300194 | 0.06 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr7_+_64838786 | 0.06 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr7_-_100061869 | 0.06 |
ENST00000332375.3
|
C7orf61
|
chromosome 7 open reading frame 61 |
| chr6_+_10528560 | 0.06 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr4_+_146402925 | 0.06 |
ENST00000302085.4
|
SMAD1
|
SMAD family member 1 |
| chr1_+_196743943 | 0.06 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr13_+_38923959 | 0.06 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr12_+_117013656 | 0.06 |
ENST00000556529.1
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2 |
| chr20_+_57226841 | 0.06 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr14_+_23344345 | 0.06 |
ENST00000551466.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr1_+_180897269 | 0.06 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr17_-_7017559 | 0.06 |
ENST00000446679.2
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chrX_-_30877837 | 0.06 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr2_-_216240386 | 0.05 |
ENST00000438981.1
|
FN1
|
fibronectin 1 |
| chr1_-_186344802 | 0.05 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr3_+_185300391 | 0.05 |
ENST00000545472.1
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr5_+_140560980 | 0.05 |
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16 |
| chr7_-_44530479 | 0.05 |
ENST00000355451.7
|
NUDCD3
|
NudC domain containing 3 |
| chr1_+_145883868 | 0.05 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr5_-_131132658 | 0.05 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr19_+_42041702 | 0.05 |
ENST00000487420.2
|
AC006129.4
|
AC006129.4 |
| chr4_-_83280767 | 0.05 |
ENST00000514671.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr19_-_39390350 | 0.05 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr1_+_17559776 | 0.05 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chrX_+_118602363 | 0.05 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr5_-_171433579 | 0.05 |
ENST00000265094.5
ENST00000393802.2 |
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr16_+_2016821 | 0.05 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chr2_-_152382500 | 0.05 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr11_-_62359027 | 0.05 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr3_+_62936098 | 0.05 |
ENST00000475886.1
ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698
|
long intergenic non-protein coding RNA 698 |
| chr12_-_11175219 | 0.05 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr1_-_24306835 | 0.05 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr20_+_21284416 | 0.05 |
ENST00000539513.1
|
XRN2
|
5'-3' exoribonuclease 2 |
| chr7_-_72972319 | 0.05 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr10_-_61513146 | 0.05 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr7_-_121784285 | 0.05 |
ENST00000417368.2
|
AASS
|
aminoadipate-semialdehyde synthase |
| chr4_+_75858290 | 0.05 |
ENST00000513238.1
|
PARM1
|
prostate androgen-regulated mucin-like protein 1 |
| chr1_+_185126598 | 0.04 |
ENST00000450350.1
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr1_-_53387352 | 0.04 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr2_-_37501692 | 0.04 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr8_+_67782984 | 0.04 |
ENST00000396592.3
ENST00000422365.2 ENST00000492775.1 |
MCMDC2
|
minichromosome maintenance domain containing 2 |
| chr5_-_159766528 | 0.04 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr7_-_45956856 | 0.04 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr12_-_92821922 | 0.04 |
ENST00000538965.1
ENST00000378487.2 |
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand |
| chr16_-_18911366 | 0.04 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_25915896 | 0.04 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr2_+_227771404 | 0.04 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr6_+_163148161 | 0.04 |
ENST00000337019.3
ENST00000366889.2 |
PACRG
|
PARK2 co-regulated |
| chr21_+_46654249 | 0.04 |
ENST00000584169.1
ENST00000328344.2 |
LINC00334
|
long intergenic non-protein coding RNA 334 |
| chr12_-_11002063 | 0.04 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr3_+_118905564 | 0.04 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr1_+_76251879 | 0.04 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.3 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.2 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.2 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.2 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.1 | GO:1904586 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) hepatocyte dedifferentiation(GO:1990828) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |