Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
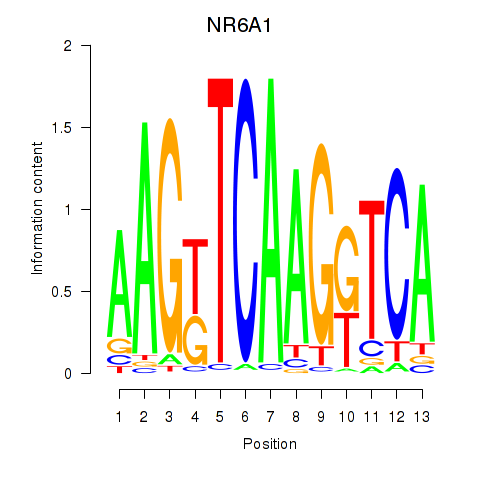
Results for NR6A1
Z-value: 0.42
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127533519_127533576 | -0.67 | 3.3e-01 | Click! |
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_64764435 | 0.96 |
ENST00000534177.1
ENST00000301887.4 |
BATF2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr19_+_10197463 | 0.47 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr2_-_231084820 | 0.35 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr9_-_38069208 | 0.27 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr8_+_74271144 | 0.25 |
ENST00000519134.1
ENST00000518355.1 |
RP11-434I12.3
|
RP11-434I12.3 |
| chr19_-_51289374 | 0.25 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr5_+_52285144 | 0.23 |
ENST00000296585.5
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| chr6_-_160166218 | 0.23 |
ENST00000537657.1
|
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr16_-_48644061 | 0.22 |
ENST00000262384.3
|
N4BP1
|
NEDD4 binding protein 1 |
| chr17_-_39093672 | 0.21 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr16_-_3422283 | 0.20 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr19_+_11670245 | 0.20 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr19_-_6720686 | 0.19 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_+_10196981 | 0.18 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr21_+_33671160 | 0.18 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr11_-_18270182 | 0.18 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr12_+_13044787 | 0.17 |
ENST00000534831.1
|
GPRC5A
|
G protein-coupled receptor, family C, group 5, member A |
| chr20_+_43803517 | 0.16 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr12_-_107168696 | 0.16 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr12_+_75874460 | 0.16 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr19_-_43383850 | 0.15 |
ENST00000436291.2
ENST00000595124.1 ENST00000244296.2 |
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr17_-_26903900 | 0.15 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr1_-_1763721 | 0.15 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr22_+_30163340 | 0.14 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr3_-_114790179 | 0.14 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_-_3306587 | 0.14 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr5_+_34757309 | 0.14 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr15_-_41166414 | 0.14 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr2_+_101437487 | 0.14 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr8_-_141645645 | 0.14 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr3_-_124560257 | 0.14 |
ENST00000496703.1
|
ITGB5
|
integrin, beta 5 |
| chr20_+_33462888 | 0.13 |
ENST00000336325.4
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr7_-_141957847 | 0.13 |
ENST00000552471.1
ENST00000547058.2 |
PRSS58
|
protease, serine, 58 |
| chrX_+_1734051 | 0.13 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr11_-_33743952 | 0.13 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr12_+_123942188 | 0.13 |
ENST00000526639.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr7_-_150777949 | 0.12 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr22_+_38609538 | 0.12 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr11_-_115127611 | 0.12 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr19_+_45542773 | 0.12 |
ENST00000544944.2
|
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr20_-_30310336 | 0.12 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr14_-_96180435 | 0.12 |
ENST00000556450.1
ENST00000555202.1 ENST00000554012.1 ENST00000402399.1 |
TCL1A
|
T-cell leukemia/lymphoma 1A |
| chr16_+_777739 | 0.12 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr9_-_130679257 | 0.12 |
ENST00000361444.3
ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr10_-_71892555 | 0.11 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr7_+_102004322 | 0.11 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr8_+_145203548 | 0.11 |
ENST00000534366.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr19_-_43586820 | 0.11 |
ENST00000406487.1
|
PSG2
|
pregnancy specific beta-1-glycoprotein 2 |
| chr7_-_43769051 | 0.11 |
ENST00000395880.3
|
COA1
|
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr15_+_76629064 | 0.11 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr5_+_34656331 | 0.11 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr8_-_87242589 | 0.11 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr19_+_41699135 | 0.11 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chrX_-_118827333 | 0.11 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr1_-_16532985 | 0.11 |
ENST00000441785.1
ENST00000449495.1 |
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr6_-_43027105 | 0.11 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr14_-_55738788 | 0.11 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr11_-_133826852 | 0.11 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr2_+_201994208 | 0.10 |
ENST00000440180.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chrX_+_1733876 | 0.10 |
ENST00000381241.3
|
ASMT
|
acetylserotonin O-methyltransferase |
| chr7_-_150777920 | 0.10 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr19_-_56709162 | 0.10 |
ENST00000589938.1
ENST00000587032.2 ENST00000586855.2 |
ZSCAN5B
|
zinc finger and SCAN domain containing 5B |
| chr11_+_110001723 | 0.10 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr10_+_24738355 | 0.10 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr6_-_31138439 | 0.10 |
ENST00000259915.8
|
POU5F1
|
POU class 5 homeobox 1 |
| chr16_-_420514 | 0.10 |
ENST00000199706.8
|
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr6_-_31620403 | 0.10 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr11_-_64512273 | 0.10 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_+_4981535 | 0.10 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr5_+_34656529 | 0.10 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr19_+_41698927 | 0.10 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr7_-_150777874 | 0.09 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr17_-_79817091 | 0.09 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr16_-_420338 | 0.09 |
ENST00000450882.1
ENST00000441883.1 ENST00000447696.1 ENST00000389675.2 |
MRPL28
|
mitochondrial ribosomal protein L28 |
| chr17_+_7338737 | 0.09 |
ENST00000323206.1
ENST00000396568.1 |
TMEM102
|
transmembrane protein 102 |
| chr3_+_46618727 | 0.09 |
ENST00000296145.5
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr19_+_41117770 | 0.09 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr10_+_47894023 | 0.09 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr6_-_31620455 | 0.09 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chr16_+_22518495 | 0.09 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr22_+_23229960 | 0.09 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr7_-_43769066 | 0.09 |
ENST00000223336.6
ENST00000310564.6 ENST00000431651.1 ENST00000415798.1 |
COA1
|
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr8_+_119294456 | 0.09 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr11_-_34379546 | 0.09 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr8_-_59412717 | 0.09 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr16_-_18801643 | 0.09 |
ENST00000322989.4
ENST00000563390.1 |
RPS15A
|
ribosomal protein S15a |
| chr5_+_34656569 | 0.09 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr11_+_65407331 | 0.08 |
ENST00000527525.1
|
SIPA1
|
signal-induced proliferation-associated 1 |
| chr11_-_111957451 | 0.08 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr16_-_18801582 | 0.08 |
ENST00000565420.1
|
RPS15A
|
ribosomal protein S15a |
| chr16_-_69448 | 0.08 |
ENST00000326592.9
|
WASH4P
|
WAS protein family homolog 4 pseudogene |
| chr19_-_43383819 | 0.08 |
ENST00000312439.6
ENST00000403380.3 |
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chrX_+_102840408 | 0.08 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr4_-_185776854 | 0.08 |
ENST00000511703.1
|
RP11-701P16.5
|
RP11-701P16.5 |
| chr17_-_57784755 | 0.08 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr1_+_6684918 | 0.08 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr3_+_39851094 | 0.08 |
ENST00000302541.6
|
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr1_+_205473720 | 0.08 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr19_-_6737241 | 0.08 |
ENST00000430424.4
ENST00000597298.1 |
GPR108
|
G protein-coupled receptor 108 |
| chr17_+_77030267 | 0.08 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr12_+_58087738 | 0.08 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr2_-_121223697 | 0.08 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr19_-_8373173 | 0.08 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr17_-_8198636 | 0.08 |
ENST00000577745.1
ENST00000579192.1 ENST00000396278.1 |
SLC25A35
|
solute carrier family 25, member 35 |
| chr17_+_48712206 | 0.08 |
ENST00000427699.1
ENST00000285238.8 |
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr12_+_54384370 | 0.08 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr13_+_23755099 | 0.08 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr11_-_72852320 | 0.07 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr6_-_31620149 | 0.07 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr14_-_91282726 | 0.07 |
ENST00000328459.6
ENST00000357056.2 |
TTC7B
|
tetratricopeptide repeat domain 7B |
| chr12_+_60058458 | 0.07 |
ENST00000548610.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_34656450 | 0.07 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr5_-_138725560 | 0.07 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr6_+_106534192 | 0.07 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr7_+_99933730 | 0.07 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr7_-_75401513 | 0.07 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chrX_+_48916497 | 0.07 |
ENST00000496529.2
ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120
|
coiled-coil domain containing 120 |
| chr2_+_219575653 | 0.07 |
ENST00000442769.1
ENST00000424644.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr5_-_176836577 | 0.07 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr6_+_149539053 | 0.07 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr10_+_106014468 | 0.07 |
ENST00000369710.4
ENST00000369713.5 ENST00000445155.1 |
GSTO1
|
glutathione S-transferase omega 1 |
| chr11_-_108464465 | 0.07 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr19_+_49999631 | 0.07 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr19_+_38865398 | 0.07 |
ENST00000585598.1
ENST00000602911.1 ENST00000592561.1 |
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr11_-_64511575 | 0.07 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr8_+_67104323 | 0.07 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr6_-_31620095 | 0.07 |
ENST00000424176.1
ENST00000456622.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr17_+_45810594 | 0.07 |
ENST00000177694.1
|
TBX21
|
T-box 21 |
| chr13_+_23755127 | 0.07 |
ENST00000545013.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr13_+_23755054 | 0.07 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr8_-_145701718 | 0.07 |
ENST00000377317.4
|
FOXH1
|
forkhead box H1 |
| chr1_+_78511586 | 0.07 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr2_-_60780546 | 0.06 |
ENST00000358510.4
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr12_+_50017184 | 0.06 |
ENST00000548825.2
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr3_-_126327398 | 0.06 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr2_-_60780536 | 0.06 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chrX_-_118827113 | 0.06 |
ENST00000394617.2
|
SEPT6
|
septin 6 |
| chr6_+_53948328 | 0.06 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr3_+_185046676 | 0.06 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr16_+_776936 | 0.06 |
ENST00000549114.1
ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr16_+_30759563 | 0.06 |
ENST00000563588.1
ENST00000565924.1 ENST00000424889.3 |
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr19_-_11669960 | 0.06 |
ENST00000589171.1
ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1
|
elongation factor 1 homolog (S. cerevisiae) |
| chr1_-_161207953 | 0.06 |
ENST00000367982.4
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_-_226111929 | 0.06 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr19_-_49956728 | 0.06 |
ENST00000601825.1
ENST00000596049.1 ENST00000599366.1 ENST00000597415.1 |
PIH1D1
|
PIH1 domain containing 1 |
| chr3_+_51976338 | 0.06 |
ENST00000417220.2
ENST00000431474.1 ENST00000398755.3 |
PARP3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr22_-_40929812 | 0.06 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr19_-_7293942 | 0.06 |
ENST00000341500.5
ENST00000302850.5 |
INSR
|
insulin receptor |
| chr16_-_3450963 | 0.06 |
ENST00000573327.1
ENST00000571906.1 ENST00000573830.1 ENST00000439568.2 ENST00000422427.2 ENST00000304926.3 ENST00000396852.4 |
ZSCAN32
|
zinc finger and SCAN domain containing 32 |
| chr9_+_132388423 | 0.06 |
ENST00000372483.4
ENST00000459968.2 ENST00000482347.1 ENST00000372481.3 |
NTMT1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr3_+_171561127 | 0.06 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr21_+_43639211 | 0.05 |
ENST00000450121.1
ENST00000398449.3 ENST00000361802.2 |
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr11_-_108464321 | 0.05 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr2_-_60780607 | 0.05 |
ENST00000537768.1
ENST00000335712.6 ENST00000356842.4 |
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr20_+_57209828 | 0.05 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr1_-_147245484 | 0.05 |
ENST00000271348.2
|
GJA5
|
gap junction protein, alpha 5, 40kDa |
| chr1_+_197886461 | 0.05 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr17_+_46970127 | 0.05 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr11_+_118754475 | 0.05 |
ENST00000292174.4
|
CXCR5
|
chemokine (C-X-C motif) receptor 5 |
| chr1_+_205473865 | 0.05 |
ENST00000506215.1
ENST00000419301.1 |
CDK18
|
cyclin-dependent kinase 18 |
| chr1_+_205473784 | 0.05 |
ENST00000478560.1
ENST00000443813.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr17_+_46970178 | 0.05 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr18_+_50278430 | 0.05 |
ENST00000578080.1
ENST00000582875.1 ENST00000412726.1 |
DCC
|
deleted in colorectal carcinoma |
| chr1_-_161102421 | 0.05 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr17_-_71410794 | 0.05 |
ENST00000424778.1
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr2_+_219575543 | 0.05 |
ENST00000457313.1
ENST00000415717.1 ENST00000392102.1 |
TTLL4
|
tubulin tyrosine ligase-like family, member 4 |
| chr7_-_137531606 | 0.05 |
ENST00000288490.5
|
DGKI
|
diacylglycerol kinase, iota |
| chr12_-_54691668 | 0.05 |
ENST00000553198.1
|
NFE2
|
nuclear factor, erythroid 2 |
| chr12_+_122242597 | 0.05 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr19_+_53761545 | 0.05 |
ENST00000341702.3
|
VN1R2
|
vomeronasal 1 receptor 2 |
| chr16_-_59788718 | 0.05 |
ENST00000564533.1
|
RP11-105C20.2
|
Uncharacterized protein |
| chr3_-_49158218 | 0.05 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr9_+_75136717 | 0.05 |
ENST00000297784.5
|
TMC1
|
transmembrane channel-like 1 |
| chr16_-_30997533 | 0.05 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr2_+_33701707 | 0.05 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_101179152 | 0.05 |
ENST00000264254.6
|
PDCL3
|
phosducin-like 3 |
| chr17_+_66509019 | 0.05 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_-_161102367 | 0.05 |
ENST00000464113.1
|
DEDD
|
death effector domain containing |
| chr16_+_22501658 | 0.05 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr16_+_30759700 | 0.05 |
ENST00000328273.7
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr17_+_48712138 | 0.05 |
ENST00000515707.1
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr18_+_616711 | 0.05 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr19_+_42381173 | 0.05 |
ENST00000221972.3
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr11_-_46141338 | 0.05 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr1_-_161208013 | 0.05 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr21_-_37838739 | 0.05 |
ENST00000399139.1
|
CLDN14
|
claudin 14 |
| chr7_+_6048856 | 0.05 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr7_-_151107767 | 0.05 |
ENST00000477459.1
|
WDR86
|
WD repeat domain 86 |
| chr22_-_21984282 | 0.05 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr17_+_40985407 | 0.05 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr17_+_73089382 | 0.05 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr11_+_63742050 | 0.05 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr18_+_616672 | 0.05 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr5_+_140027355 | 0.04 |
ENST00000417647.2
ENST00000507593.1 ENST00000508301.1 |
IK
|
IK cytokine, down-regulator of HLA II |
| chr6_+_46761118 | 0.04 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr3_+_57882061 | 0.04 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr22_+_18593507 | 0.04 |
ENST00000330423.3
|
TUBA8
|
tubulin, alpha 8 |
| chr9_+_132096166 | 0.04 |
ENST00000436710.1
|
RP11-65J3.1
|
RP11-65J3.1 |
| chrY_-_21906623 | 0.04 |
ENST00000382806.2
|
KDM5D
|
lysine (K)-specific demethylase 5D |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.2 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.3 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0090625 | siRNA loading onto RISC involved in RNA interference(GO:0035087) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0035570 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.0 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0098904 | cell communication by chemical coupling(GO:0010643) SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.0 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.0 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |