Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
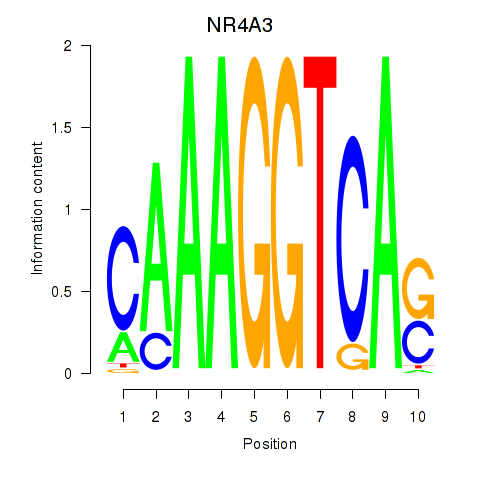
Results for NR4A3
Z-value: 0.41
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | nuclear receptor subfamily 4 group A member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | -0.88 | 1.2e-01 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_150563 | 0.33 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr13_+_113760098 | 0.27 |
ENST00000346342.3
ENST00000541084.1 ENST00000375581.3 |
F7
|
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr19_+_38880695 | 0.24 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr17_+_4675175 | 0.21 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr11_-_73720122 | 0.19 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr22_+_39493207 | 0.19 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr12_-_53207842 | 0.18 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr8_-_131028782 | 0.17 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr12_+_57854274 | 0.17 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr17_-_64216748 | 0.15 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr20_-_45976816 | 0.14 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_32655110 | 0.13 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_-_48867171 | 0.13 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr17_-_2318731 | 0.12 |
ENST00000609667.1
|
AC006435.1
|
Uncharacterized protein |
| chr19_-_48867291 | 0.12 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr21_-_27107198 | 0.11 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_+_155484155 | 0.11 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr21_-_27107283 | 0.11 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_+_24645807 | 0.10 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_+_92219919 | 0.10 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr16_-_30538079 | 0.10 |
ENST00000562803.1
|
ZNF768
|
zinc finger protein 768 |
| chr17_-_79167828 | 0.10 |
ENST00000570817.1
|
AZI1
|
5-azacytidine induced 1 |
| chr5_-_1801408 | 0.10 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr10_+_102729249 | 0.10 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr17_-_77967433 | 0.10 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_12679171 | 0.10 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr16_-_66952742 | 0.10 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr14_-_107283278 | 0.10 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr10_-_75351088 | 0.09 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr1_+_24645865 | 0.09 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_210502238 | 0.09 |
ENST00000545154.1
ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT
|
hedgehog acyltransferase |
| chr1_+_24646002 | 0.09 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_-_89878369 | 0.09 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr19_-_39303576 | 0.09 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr21_-_27107344 | 0.09 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr1_-_115301235 | 0.09 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr20_+_19738792 | 0.09 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr20_-_46415297 | 0.09 |
ENST00000467815.1
ENST00000359930.4 |
SULF2
|
sulfatase 2 |
| chr9_+_135937365 | 0.09 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr10_-_8095412 | 0.09 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chr1_-_1356719 | 0.09 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr12_+_57828521 | 0.09 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr10_-_30638090 | 0.09 |
ENST00000421701.1
ENST00000263063.4 |
MTPAP
|
mitochondrial poly(A) polymerase |
| chr17_-_6616678 | 0.09 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr7_-_37956409 | 0.08 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr16_+_28874345 | 0.08 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr16_-_10652993 | 0.08 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr9_+_130890612 | 0.08 |
ENST00000443493.1
|
AL590708.2
|
AL590708.2 |
| chr9_-_75567962 | 0.08 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr19_-_51289436 | 0.08 |
ENST00000562076.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr18_+_12947126 | 0.08 |
ENST00000592170.1
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr14_+_32546274 | 0.08 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr12_+_49717019 | 0.08 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr16_+_28875126 | 0.08 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr20_-_46415341 | 0.08 |
ENST00000484875.1
ENST00000361612.4 |
SULF2
|
sulfatase 2 |
| chr1_+_24645915 | 0.08 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_235116495 | 0.08 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr19_-_4535233 | 0.08 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr4_-_111563076 | 0.08 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr17_-_7081435 | 0.07 |
ENST00000380920.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr16_+_88872176 | 0.07 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr3_-_47484661 | 0.07 |
ENST00000495603.2
|
SCAP
|
SREBF chaperone |
| chr14_-_23791484 | 0.07 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr16_+_2880254 | 0.07 |
ENST00000570670.1
|
ZG16B
|
zymogen granule protein 16B |
| chr16_+_81040103 | 0.07 |
ENST00000305850.5
ENST00000299572.5 |
CENPN
|
centromere protein N |
| chr11_-_64013663 | 0.07 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chrX_-_154689596 | 0.07 |
ENST00000369444.2
|
H2AFB3
|
H2A histone family, member B3 |
| chr8_+_99956662 | 0.07 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr15_-_55790515 | 0.07 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr10_+_101542462 | 0.07 |
ENST00000370449.4
ENST00000370434.1 |
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr5_-_107703556 | 0.07 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr10_-_76995675 | 0.07 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr14_+_37667118 | 0.07 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr5_+_111964133 | 0.07 |
ENST00000508879.1
ENST00000507565.1 |
RP11-159K7.2
|
RP11-159K7.2 |
| chr12_-_106697974 | 0.07 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr7_-_87104963 | 0.07 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr19_+_19639670 | 0.06 |
ENST00000436027.5
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr1_-_20306909 | 0.06 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr1_+_16062820 | 0.06 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr15_-_45422056 | 0.06 |
ENST00000267803.4
ENST00000559014.1 ENST00000558851.1 ENST00000559988.1 ENST00000558996.1 ENST00000558422.1 ENST00000559226.1 ENST00000558326.1 ENST00000558377.1 ENST00000559644.1 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr1_-_211848899 | 0.06 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr18_-_3247084 | 0.06 |
ENST00000609924.1
|
RP13-270P17.3
|
RP13-270P17.3 |
| chr13_+_36920569 | 0.06 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr16_-_2168079 | 0.06 |
ENST00000488185.2
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr14_-_81408063 | 0.06 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr3_+_172034218 | 0.06 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr12_+_12223867 | 0.06 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr1_-_1356628 | 0.06 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr1_-_40042416 | 0.06 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr17_+_79373540 | 0.06 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr1_+_163291680 | 0.06 |
ENST00000450453.2
ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2
|
NUF2, NDC80 kinetochore complex component |
| chr3_-_9291063 | 0.06 |
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr17_-_46682321 | 0.06 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr19_-_15236470 | 0.06 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr4_+_57253672 | 0.06 |
ENST00000602927.1
|
RP11-646I6.5
|
RP11-646I6.5 |
| chr6_-_34360413 | 0.06 |
ENST00000607016.1
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr22_+_31518938 | 0.06 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr18_+_21719018 | 0.06 |
ENST00000585037.1
ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr7_-_98805129 | 0.06 |
ENST00000327442.6
|
KPNA7
|
karyopherin alpha 7 (importin alpha 8) |
| chr1_+_45205591 | 0.06 |
ENST00000455186.1
|
KIF2C
|
kinesin family member 2C |
| chr9_-_130487143 | 0.05 |
ENST00000419060.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr5_-_64858944 | 0.05 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr22_+_50925213 | 0.05 |
ENST00000395733.3
ENST00000216075.6 ENST00000395732.3 |
MIOX
|
myo-inositol oxygenase |
| chr17_+_7211656 | 0.05 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_+_10068095 | 0.05 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr11_-_117698765 | 0.05 |
ENST00000532119.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr7_-_27169801 | 0.05 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr16_-_30537839 | 0.05 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr19_-_57967854 | 0.05 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr6_-_11807277 | 0.05 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr19_-_18995029 | 0.05 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr11_-_790060 | 0.05 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_+_165600436 | 0.05 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr11_-_118272610 | 0.05 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr19_+_41949054 | 0.05 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chr1_+_1217305 | 0.05 |
ENST00000470022.1
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr18_+_56530794 | 0.05 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr5_-_95158644 | 0.05 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr12_+_56624436 | 0.05 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr14_+_35451880 | 0.05 |
ENST00000554803.1
ENST00000555746.1 |
SRP54
|
signal recognition particle 54kDa |
| chr8_+_99956759 | 0.05 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr17_+_79679369 | 0.05 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr19_-_3062881 | 0.05 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr3_+_133292851 | 0.05 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr14_-_24729251 | 0.05 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr8_+_11534462 | 0.05 |
ENST00000528712.1
ENST00000532977.1 |
GATA4
|
GATA binding protein 4 |
| chr11_+_118272328 | 0.05 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr14_+_100532771 | 0.05 |
ENST00000557153.1
|
EVL
|
Enah/Vasp-like |
| chr1_+_184020830 | 0.05 |
ENST00000533373.1
ENST00000423085.2 |
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr2_+_121103706 | 0.05 |
ENST00000295228.3
|
INHBB
|
inhibin, beta B |
| chr13_-_46679144 | 0.05 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr11_-_64703354 | 0.05 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr3_-_46000146 | 0.05 |
ENST00000438446.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr19_-_49552363 | 0.05 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr22_+_39493268 | 0.05 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr19_-_15236562 | 0.05 |
ENST00000263383.3
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr6_-_53013620 | 0.05 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr10_+_116853091 | 0.05 |
ENST00000526946.1
|
ATRNL1
|
attractin-like 1 |
| chr20_+_13202418 | 0.05 |
ENST00000262487.4
|
ISM1
|
isthmin 1, angiogenesis inhibitor |
| chr7_-_111202511 | 0.05 |
ENST00000452895.1
ENST00000452753.1 ENST00000331762.3 |
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_+_13141010 | 0.05 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr17_-_73178599 | 0.05 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr13_+_49822041 | 0.05 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_-_141703713 | 0.05 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr3_-_197300194 | 0.05 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr3_+_184055240 | 0.05 |
ENST00000383847.2
|
FAM131A
|
family with sequence similarity 131, member A |
| chr5_-_179718929 | 0.05 |
ENST00000425491.2
|
MAPK9
|
mitogen-activated protein kinase 9 |
| chr11_-_8832182 | 0.05 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr11_-_57282349 | 0.05 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr19_-_29218538 | 0.04 |
ENST00000592347.1
ENST00000586528.1 |
AC005307.3
|
AC005307.3 |
| chr1_+_203096831 | 0.04 |
ENST00000337894.4
|
ADORA1
|
adenosine A1 receptor |
| chr2_+_28974603 | 0.04 |
ENST00000441461.1
ENST00000358506.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr11_+_102552041 | 0.04 |
ENST00000537079.1
|
RP11-817J15.3
|
Uncharacterized protein |
| chr17_+_79679299 | 0.04 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr10_-_100206642 | 0.04 |
ENST00000361490.4
ENST00000338546.5 ENST00000325103.6 |
HPS1
|
Hermansky-Pudlak syndrome 1 |
| chr11_-_8832521 | 0.04 |
ENST00000530438.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr1_-_27701307 | 0.04 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr1_+_119957554 | 0.04 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr11_-_66445219 | 0.04 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr11_+_278365 | 0.04 |
ENST00000534750.1
|
NLRP6
|
NLR family, pyrin domain containing 6 |
| chr17_-_8066843 | 0.04 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr11_-_117698787 | 0.04 |
ENST00000260287.2
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr1_-_45253377 | 0.04 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr16_-_28937027 | 0.04 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_+_67563250 | 0.04 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr2_+_242167319 | 0.04 |
ENST00000601871.1
|
AC104841.2
|
HCG1777198, isoform CRA_a; PRO2900; Uncharacterized protein |
| chr9_+_139746792 | 0.04 |
ENST00000317446.2
ENST00000445819.1 |
MAMDC4
|
MAM domain containing 4 |
| chr10_-_76995769 | 0.04 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr7_-_17500294 | 0.04 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr16_+_2076869 | 0.04 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr11_-_117699413 | 0.04 |
ENST00000528014.1
|
FXYD2
|
FXYD domain containing ion transport regulator 2 |
| chr2_+_128175997 | 0.04 |
ENST00000234071.3
ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr16_+_691792 | 0.04 |
ENST00000307650.4
|
FAM195A
|
family with sequence similarity 195, member A |
| chr16_-_58328923 | 0.04 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr17_-_49021974 | 0.04 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr12_+_58013693 | 0.04 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr5_+_68462837 | 0.04 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr6_+_108616054 | 0.04 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr7_-_229557 | 0.04 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr22_+_30115986 | 0.04 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr2_+_28974668 | 0.04 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_164534657 | 0.04 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr12_+_53443963 | 0.04 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr17_-_73179046 | 0.04 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr7_+_121513143 | 0.04 |
ENST00000393386.2
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr3_-_150690786 | 0.04 |
ENST00000327047.1
|
CLRN1
|
clarin 1 |
| chr11_+_66624527 | 0.04 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_+_1248547 | 0.04 |
ENST00000586757.1
ENST00000300952.2 |
MIDN
|
midnolin |
| chr22_+_46546494 | 0.04 |
ENST00000396000.2
ENST00000262735.5 ENST00000420804.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr22_-_24110063 | 0.04 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr2_-_231989808 | 0.04 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr6_+_31637944 | 0.04 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr10_+_14880157 | 0.04 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr14_+_23791159 | 0.04 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr4_+_124317940 | 0.04 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr1_+_45274154 | 0.04 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr6_-_43197189 | 0.04 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr5_-_127674883 | 0.04 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr12_+_124457670 | 0.04 |
ENST00000539644.1
|
ZNF664
|
zinc finger protein 664 |
| chr12_-_2966193 | 0.04 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.1 | GO:0071422 | succinate transport(GO:0015744) tricarboxylic acid transmembrane transport(GO:0035674) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.0 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.0 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.0 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.0 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.0 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |