Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
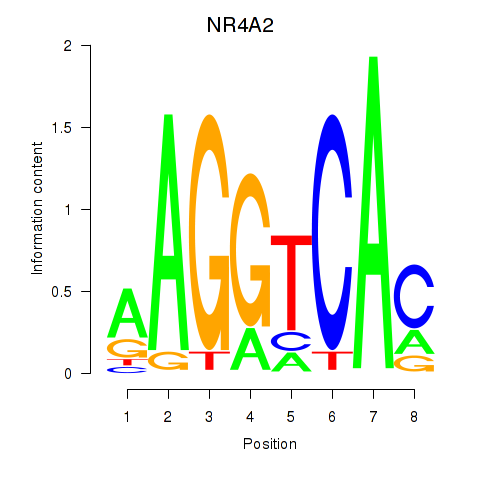
Results for NR4A2
Z-value: 0.48
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157189180_157189290 | -0.68 | 3.2e-01 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_51289374 | 0.53 |
ENST00000563228.1
|
CTD-2568A17.1
|
CTD-2568A17.1 |
| chr10_+_81065975 | 0.44 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr17_-_42988004 | 0.42 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr3_-_117716418 | 0.40 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr17_-_1553346 | 0.34 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr17_+_7533439 | 0.30 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr14_+_21492331 | 0.29 |
ENST00000533984.1
ENST00000532213.2 |
AL161668.5
|
AL161668.5 |
| chr12_-_56753858 | 0.28 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
| chr22_-_39239987 | 0.26 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr22_+_46546406 | 0.26 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chrX_-_21776281 | 0.25 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr22_+_32149927 | 0.25 |
ENST00000437411.1
ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5
|
DEP domain containing 5 |
| chr20_+_43343886 | 0.24 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr16_-_30997533 | 0.22 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr19_+_35629702 | 0.22 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr22_+_37959647 | 0.21 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr20_+_44421137 | 0.20 |
ENST00000415790.1
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr7_+_129007964 | 0.20 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr10_-_128359074 | 0.19 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr22_+_51176624 | 0.19 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr12_-_16760195 | 0.18 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr20_+_44462749 | 0.18 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr10_-_94301107 | 0.18 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr6_-_34664612 | 0.17 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr18_-_43678241 | 0.17 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr12_+_120875910 | 0.16 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr9_-_136344237 | 0.16 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr9_+_112852477 | 0.16 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr20_+_3776371 | 0.16 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr5_-_173217931 | 0.16 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr9_-_136344197 | 0.15 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr4_+_41614720 | 0.15 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_+_1407733 | 0.15 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr22_+_30821732 | 0.15 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr2_+_228678550 | 0.15 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr2_-_27712583 | 0.15 |
ENST00000260570.3
ENST00000359466.6 ENST00000416524.2 |
IFT172
|
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr7_-_127671674 | 0.15 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr20_+_58179582 | 0.14 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chr1_+_155179012 | 0.14 |
ENST00000609421.1
|
MTX1
|
metaxin 1 |
| chr16_+_28874860 | 0.14 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr19_+_589893 | 0.14 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr12_+_7169887 | 0.14 |
ENST00000542978.1
|
C1S
|
complement component 1, s subcomponent |
| chr5_-_149792295 | 0.14 |
ENST00000518797.1
ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74
|
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr5_-_176981417 | 0.14 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr2_-_207023918 | 0.14 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr3_-_49459865 | 0.14 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr11_-_64512273 | 0.13 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_+_18121356 | 0.13 |
ENST00000317582.5
ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr17_+_79670386 | 0.13 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr12_+_93963590 | 0.13 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr20_-_48532019 | 0.13 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr22_+_30792980 | 0.13 |
ENST00000403484.1
ENST00000405717.3 ENST00000402592.3 |
SEC14L2
|
SEC14-like 2 (S. cerevisiae) |
| chr17_+_73975292 | 0.13 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr11_+_77899842 | 0.13 |
ENST00000530267.1
|
USP35
|
ubiquitin specific peptidase 35 |
| chr16_+_11038345 | 0.13 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr16_-_69368774 | 0.13 |
ENST00000562949.1
|
RP11-343C2.12
|
Conserved oligomeric Golgi complex subunit 8 |
| chr15_-_41120896 | 0.13 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr12_-_46121554 | 0.12 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
| chr17_-_49021974 | 0.12 |
ENST00000501718.2
|
RP11-700H6.1
|
RP11-700H6.1 |
| chr2_+_220299547 | 0.12 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr8_+_145582633 | 0.12 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr7_+_39663485 | 0.12 |
ENST00000436179.1
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr1_-_229569834 | 0.12 |
ENST00000366684.3
ENST00000366683.2 |
ACTA1
|
actin, alpha 1, skeletal muscle |
| chr5_+_218356 | 0.12 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr15_-_55562479 | 0.12 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr16_-_4466565 | 0.12 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr7_-_127672146 | 0.11 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr11_+_67798114 | 0.11 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr20_+_3776936 | 0.11 |
ENST00000439880.2
|
CDC25B
|
cell division cycle 25B |
| chr1_-_201140673 | 0.11 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr11_-_64511789 | 0.11 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_-_207024134 | 0.11 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chrX_+_135230712 | 0.11 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr22_-_30783075 | 0.11 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr17_+_48423453 | 0.10 |
ENST00000017003.2
ENST00000509778.1 ENST00000507602.1 |
XYLT2
|
xylosyltransferase II |
| chr1_-_116311402 | 0.10 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr7_-_150777949 | 0.10 |
ENST00000482571.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr8_-_100905850 | 0.10 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_-_220110111 | 0.10 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr15_-_41408339 | 0.10 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr1_+_206858328 | 0.10 |
ENST00000367103.3
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr12_+_120875887 | 0.10 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr7_-_151433393 | 0.10 |
ENST00000492843.1
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_45140400 | 0.10 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr7_-_150777920 | 0.10 |
ENST00000353841.2
ENST00000297532.6 |
FASTK
|
Fas-activated serine/threonine kinase |
| chr7_-_151433342 | 0.10 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_-_109683446 | 0.10 |
ENST00000372057.1
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr1_-_27816556 | 0.10 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr12_-_120554622 | 0.09 |
ENST00000229340.5
|
RAB35
|
RAB35, member RAS oncogene family |
| chr17_-_2169425 | 0.09 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr11_+_122709200 | 0.09 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr11_+_67798363 | 0.09 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr2_-_26467557 | 0.09 |
ENST00000380649.3
|
HADHA
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr10_+_81107216 | 0.09 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr9_+_115983808 | 0.09 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr2_+_85804614 | 0.09 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr8_-_100905363 | 0.09 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr12_+_50794891 | 0.09 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr12_-_120554534 | 0.09 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr12_+_6309963 | 0.09 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr22_+_30163340 | 0.09 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr2_+_210288760 | 0.08 |
ENST00000199940.6
|
MAP2
|
microtubule-associated protein 2 |
| chr19_-_49552363 | 0.08 |
ENST00000448456.3
ENST00000355414.2 |
CGB8
|
chorionic gonadotropin, beta polypeptide 8 |
| chr14_-_48144157 | 0.08 |
ENST00000399232.2
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr17_+_19281787 | 0.08 |
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7 |
| chr16_-_68269971 | 0.08 |
ENST00000565858.1
|
ESRP2
|
epithelial splicing regulatory protein 2 |
| chr1_-_154164534 | 0.08 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr18_-_12377001 | 0.08 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chrX_+_44732757 | 0.08 |
ENST00000377967.4
ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A
|
lysine (K)-specific demethylase 6A |
| chr12_+_119616447 | 0.08 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr7_+_45613958 | 0.08 |
ENST00000297323.7
|
ADCY1
|
adenylate cyclase 1 (brain) |
| chr12_-_16760021 | 0.08 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_2614927 | 0.08 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr1_+_36690011 | 0.07 |
ENST00000354618.5
ENST00000469141.2 ENST00000478853.1 |
THRAP3
|
thyroid hormone receptor associated protein 3 |
| chr15_-_41408409 | 0.07 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr1_-_104239302 | 0.07 |
ENST00000446703.1
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr2_+_119981384 | 0.07 |
ENST00000393108.2
ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3
|
STEAP family member 3, metalloreductase |
| chr20_+_13765596 | 0.07 |
ENST00000378106.5
ENST00000463598.1 |
NDUFAF5
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
| chr11_-_64511575 | 0.07 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr15_+_75335604 | 0.07 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr8_-_93115445 | 0.07 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_+_22605304 | 0.07 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr7_-_150777874 | 0.07 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr11_+_77899920 | 0.07 |
ENST00000528910.1
ENST00000529308.1 |
USP35
|
ubiquitin specific peptidase 35 |
| chr6_+_139456226 | 0.06 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr2_-_121223697 | 0.06 |
ENST00000593290.1
|
FLJ14816
|
long intergenic non-protein coding RNA 1101 |
| chr19_-_2328572 | 0.06 |
ENST00000252622.10
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr20_+_37590942 | 0.06 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr4_+_81118647 | 0.06 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr19_-_46476791 | 0.06 |
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2 |
| chr4_+_146560245 | 0.06 |
ENST00000541599.1
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_20255123 | 0.06 |
ENST00000504154.1
ENST00000273739.5 |
SLIT2
|
slit homolog 2 (Drosophila) |
| chr17_-_2615031 | 0.06 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr15_-_42783303 | 0.06 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr8_+_145149930 | 0.06 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr4_+_26321284 | 0.06 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_-_23395623 | 0.06 |
ENST00000556043.1
|
PRMT5
|
protein arginine methyltransferase 5 |
| chr17_-_7531121 | 0.06 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr11_-_63993690 | 0.06 |
ENST00000394546.2
ENST00000541278.1 |
TRPT1
|
tRNA phosphotransferase 1 |
| chr5_-_139937895 | 0.06 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr7_-_105029812 | 0.06 |
ENST00000482897.1
|
SRPK2
|
SRSF protein kinase 2 |
| chr7_-_107642348 | 0.06 |
ENST00000393561.1
|
LAMB1
|
laminin, beta 1 |
| chr14_-_61191049 | 0.06 |
ENST00000556952.3
|
SIX4
|
SIX homeobox 4 |
| chr5_-_149829244 | 0.06 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr12_+_58013693 | 0.05 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr2_-_176046391 | 0.05 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr3_-_150966902 | 0.05 |
ENST00000424796.2
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr4_-_151936865 | 0.05 |
ENST00000535741.1
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr4_-_65275162 | 0.05 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chr10_+_51371390 | 0.05 |
ENST00000478381.1
ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr3_-_49158218 | 0.05 |
ENST00000417901.1
ENST00000306026.5 ENST00000434032.2 |
USP19
|
ubiquitin specific peptidase 19 |
| chr2_+_234686976 | 0.05 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr19_-_46272106 | 0.05 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr5_+_66124590 | 0.05 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr22_+_18121562 | 0.05 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr3_-_49459878 | 0.05 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr19_+_49547099 | 0.05 |
ENST00000301408.6
|
CGB5
|
chorionic gonadotropin, beta polypeptide 5 |
| chr8_-_70745575 | 0.05 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chrY_-_15591818 | 0.05 |
ENST00000382893.1
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr19_+_46850251 | 0.05 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr16_+_3704822 | 0.05 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr22_-_24110063 | 0.05 |
ENST00000520222.1
ENST00000401675.3 |
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr14_-_21492113 | 0.05 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr19_+_4153598 | 0.05 |
ENST00000078445.2
ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3
|
cAMP responsive element binding protein 3-like 3 |
| chr5_-_149829314 | 0.05 |
ENST00000407193.1
|
RPS14
|
ribosomal protein S14 |
| chr1_-_234667504 | 0.05 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr15_-_43882140 | 0.05 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr5_-_149829294 | 0.05 |
ENST00000401695.3
|
RPS14
|
ribosomal protein S14 |
| chr7_-_81399438 | 0.05 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_-_17380630 | 0.04 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr8_+_134125727 | 0.04 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chrX_+_153639856 | 0.04 |
ENST00000426834.1
ENST00000369790.4 ENST00000454722.1 ENST00000350743.4 ENST00000299328.5 ENST00000351413.4 |
TAZ
|
tafazzin |
| chr12_+_51818555 | 0.04 |
ENST00000453097.2
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr3_+_159557637 | 0.04 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr9_+_129567282 | 0.04 |
ENST00000449886.1
ENST00000373464.4 ENST00000450858.1 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr10_+_80828774 | 0.04 |
ENST00000334512.5
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr17_+_36508826 | 0.04 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr12_-_57039739 | 0.04 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr1_+_201924619 | 0.04 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr1_-_20987851 | 0.04 |
ENST00000464364.1
ENST00000602624.2 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr20_-_60718430 | 0.04 |
ENST00000370873.4
ENST00000370858.3 |
PSMA7
|
proteasome (prosome, macropain) subunit, alpha type, 7 |
| chr3_+_128968437 | 0.04 |
ENST00000314797.6
|
COPG1
|
coatomer protein complex, subunit gamma 1 |
| chr20_-_39946237 | 0.04 |
ENST00000441102.2
ENST00000559234.1 |
ZHX3
|
zinc fingers and homeoboxes 3 |
| chr2_-_207024233 | 0.04 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr8_-_100905925 | 0.04 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr1_-_20987889 | 0.04 |
ENST00000415136.2
|
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr17_-_9940058 | 0.04 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chr7_+_117864708 | 0.04 |
ENST00000357099.4
ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7
|
ankyrin repeat domain 7 |
| chr22_+_40573921 | 0.04 |
ENST00000454349.2
ENST00000335727.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr22_-_27620603 | 0.04 |
ENST00000418271.1
ENST00000444114.1 |
RP5-1172A22.1
|
RP5-1172A22.1 |
| chr11_+_67007518 | 0.04 |
ENST00000530342.1
ENST00000308783.5 |
KDM2A
|
lysine (K)-specific demethylase 2A |
| chr2_+_220491973 | 0.04 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr6_+_36683256 | 0.04 |
ENST00000229824.8
|
RAB44
|
RAB44, member RAS oncogene family |
| chr15_+_78441663 | 0.04 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr6_+_90272488 | 0.03 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr17_+_45973516 | 0.03 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr4_-_140216948 | 0.03 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_+_36858694 | 0.03 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr20_-_33999766 | 0.03 |
ENST00000349714.5
ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr9_-_138853156 | 0.03 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0045360 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.0 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.2 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.3 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070470 | plasma membrane respiratory chain(GO:0070470) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.0 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |