Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
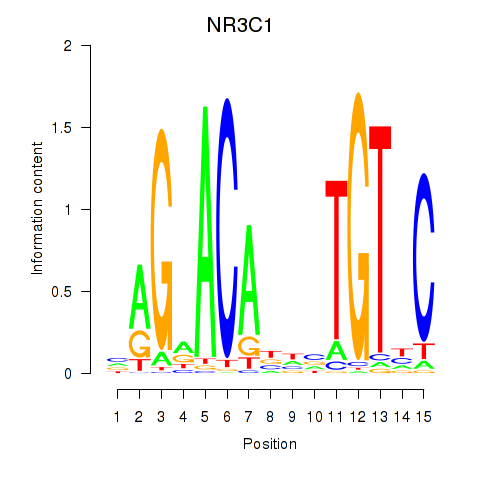
Results for NR3C1
Z-value: 0.43
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | nuclear receptor subfamily 3 group C member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142784003_142784065 | -0.91 | 8.9e-02 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_145060593 | 0.55 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr2_-_191878162 | 0.47 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr16_+_56716336 | 0.39 |
ENST00000394485.4
ENST00000562939.1 |
MT1X
|
metallothionein 1X |
| chr11_-_102668879 | 0.38 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr16_+_56642041 | 0.36 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr16_+_56642489 | 0.31 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr11_+_5711010 | 0.28 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr4_+_15779901 | 0.25 |
ENST00000226279.3
|
CD38
|
CD38 molecule |
| chr1_-_27998689 | 0.23 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr11_+_5710919 | 0.23 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr21_-_47352477 | 0.21 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr17_+_74380683 | 0.20 |
ENST00000592299.1
ENST00000590959.1 ENST00000323374.4 |
SPHK1
|
sphingosine kinase 1 |
| chr19_-_30199516 | 0.18 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr7_-_139727118 | 0.18 |
ENST00000484111.1
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr7_-_134143841 | 0.17 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr14_-_61124977 | 0.17 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr1_-_27816556 | 0.17 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr5_+_135383008 | 0.17 |
ENST00000508767.1
ENST00000604555.1 |
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr11_+_57365150 | 0.16 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr20_-_47894569 | 0.16 |
ENST00000371744.1
ENST00000371752.1 ENST00000396105.1 |
ZNFX1
|
zinc finger, NFX1-type containing 1 |
| chr17_+_74381343 | 0.16 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr19_+_4229495 | 0.16 |
ENST00000221847.5
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr18_+_21464737 | 0.15 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chr6_+_29795595 | 0.15 |
ENST00000360323.6
ENST00000376818.3 ENST00000376815.3 |
HLA-G
|
major histocompatibility complex, class I, G |
| chr3_+_38179969 | 0.14 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr8_+_128427857 | 0.14 |
ENST00000391675.1
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr5_-_146461027 | 0.14 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_-_93519471 | 0.14 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr22_-_36635225 | 0.14 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chr6_+_32811885 | 0.14 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr10_+_90562705 | 0.13 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr8_-_23712312 | 0.13 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr8_-_23282797 | 0.13 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr6_+_127898312 | 0.13 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr11_-_627143 | 0.13 |
ENST00000176195.3
|
SCT
|
secretin |
| chr19_+_57874835 | 0.13 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr10_+_99627889 | 0.12 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr10_-_90967063 | 0.12 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr1_+_19967014 | 0.12 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr3_+_98699880 | 0.12 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr19_-_53426700 | 0.12 |
ENST00000596623.1
|
ZNF888
|
zinc finger protein 888 |
| chr14_+_24641062 | 0.12 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr14_-_24616426 | 0.12 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr6_+_32811861 | 0.12 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr2_+_208104351 | 0.12 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr1_+_6511651 | 0.11 |
ENST00000434576.1
|
ESPN
|
espin |
| chr2_+_208104497 | 0.11 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr12_-_96389702 | 0.11 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr17_-_18585131 | 0.11 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr12_-_7245125 | 0.11 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr6_+_32821924 | 0.11 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_+_24641820 | 0.11 |
ENST00000560501.1
|
REC8
|
REC8 meiotic recombination protein |
| chr11_+_65851443 | 0.11 |
ENST00000533756.1
|
PACS1
|
phosphofurin acidic cluster sorting protein 1 |
| chr17_-_15496722 | 0.11 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr7_-_75241096 | 0.11 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr1_+_41448820 | 0.11 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr6_-_32821599 | 0.11 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_+_75311019 | 0.11 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr12_-_7244469 | 0.11 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr4_+_75310851 | 0.11 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr12_-_7245152 | 0.11 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr18_+_34124507 | 0.11 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr22_+_31277661 | 0.11 |
ENST00000454145.1
ENST00000453621.1 ENST00000431368.1 ENST00000535268.1 |
OSBP2
|
oxysterol binding protein 2 |
| chr2_-_31637560 | 0.10 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr17_+_36905613 | 0.10 |
ENST00000539023.1
|
CTB-58E17.5
|
Uncharacterized protein; cDNA FLJ52623 |
| chr12_-_7245018 | 0.10 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chrX_+_71401570 | 0.10 |
ENST00000496835.2
ENST00000446576.1 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr1_-_45965525 | 0.10 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr22_+_50981079 | 0.10 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr17_-_8027402 | 0.10 |
ENST00000541682.2
ENST00000317814.4 ENST00000577735.1 |
HES7
|
hes family bHLH transcription factor 7 |
| chr16_+_57438679 | 0.10 |
ENST00000219244.4
|
CCL17
|
chemokine (C-C motif) ligand 17 |
| chr20_-_36794938 | 0.10 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr14_-_77542485 | 0.10 |
ENST00000556781.1
ENST00000557526.1 ENST00000555512.1 |
RP11-7F17.3
|
RP11-7F17.3 |
| chr9_-_107690420 | 0.10 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr13_-_41111323 | 0.10 |
ENST00000595486.1
|
AL133318.1
|
Uncharacterized protein |
| chr10_+_90562487 | 0.10 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr6_-_31324943 | 0.10 |
ENST00000412585.2
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr13_+_21714653 | 0.10 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr8_-_7320974 | 0.10 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr11_-_104916034 | 0.10 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr14_+_101295948 | 0.09 |
ENST00000452514.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_-_61646054 | 0.09 |
ENST00000527379.1
|
FADS3
|
fatty acid desaturase 3 |
| chr9_-_19127474 | 0.09 |
ENST00000380465.3
ENST00000380464.3 ENST00000411567.1 ENST00000276914.2 |
PLIN2
|
perilipin 2 |
| chr15_+_67430339 | 0.09 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr14_+_24616588 | 0.09 |
ENST00000324103.6
ENST00000559260.1 |
RNF31
|
ring finger protein 31 |
| chr16_-_74734742 | 0.09 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr10_+_70847852 | 0.09 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr17_+_4843352 | 0.09 |
ENST00000573404.1
ENST00000576452.1 |
RNF167
|
ring finger protein 167 |
| chr13_+_111748183 | 0.09 |
ENST00000422994.1
|
LINC00368
|
long intergenic non-protein coding RNA 368 |
| chr4_+_89444961 | 0.09 |
ENST00000513325.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr7_+_871559 | 0.09 |
ENST00000421580.1
|
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr2_+_10560147 | 0.09 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chr19_-_47128294 | 0.09 |
ENST00000596260.1
ENST00000597185.1 ENST00000598865.1 ENST00000594275.1 ENST00000291294.2 |
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP) |
| chr1_-_145715565 | 0.09 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr6_-_160114260 | 0.09 |
ENST00000367054.2
ENST00000367055.4 ENST00000444946.2 ENST00000452684.2 |
SOD2
|
superoxide dismutase 2, mitochondrial |
| chr5_+_49961727 | 0.09 |
ENST00000505697.2
ENST00000503750.2 ENST00000514342.2 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_-_27235447 | 0.09 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr8_+_7705398 | 0.09 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr22_+_38609538 | 0.08 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr12_+_93115281 | 0.08 |
ENST00000549856.1
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr20_-_56285595 | 0.08 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_+_31950150 | 0.08 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr1_-_47069886 | 0.08 |
ENST00000371946.4
ENST00000371945.4 ENST00000428112.2 ENST00000529170.1 |
MKNK1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr9_+_131683174 | 0.08 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_+_44514040 | 0.08 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr12_-_7245080 | 0.08 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr4_-_140222358 | 0.08 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_+_228395755 | 0.08 |
ENST00000284548.11
ENST00000570156.2 ENST00000422127.1 ENST00000366707.4 ENST00000366709.4 |
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr15_+_41099788 | 0.08 |
ENST00000299173.10
ENST00000566407.1 |
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr19_-_55677920 | 0.08 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr5_-_58571935 | 0.08 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_35630926 | 0.08 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_+_52626898 | 0.08 |
ENST00000331817.5
|
KRT7
|
keratin 7 |
| chr11_+_18287721 | 0.08 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr11_-_7698453 | 0.08 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr19_-_37958318 | 0.08 |
ENST00000316950.6
ENST00000591710.1 |
ZNF569
|
zinc finger protein 569 |
| chr6_+_32006159 | 0.07 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr15_+_90728145 | 0.07 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr14_-_62217779 | 0.07 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr9_+_126777676 | 0.07 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr17_+_68047418 | 0.07 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr19_+_46498704 | 0.07 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr17_-_38978847 | 0.07 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr1_-_1149506 | 0.07 |
ENST00000379236.3
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4 |
| chr6_-_133035185 | 0.07 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr14_-_88459182 | 0.07 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr20_-_36794902 | 0.07 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr3_+_9944303 | 0.07 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr3_-_171489085 | 0.07 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr16_+_30751953 | 0.07 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr6_-_27835357 | 0.07 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr1_+_207262881 | 0.07 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr11_-_3078838 | 0.07 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr1_-_53833812 | 0.07 |
ENST00000449958.1
|
RP11-117D22.2
|
RP11-117D22.2 |
| chr7_+_55086794 | 0.07 |
ENST00000275493.2
ENST00000442591.1 |
EGFR
|
epidermal growth factor receptor |
| chr2_+_201987200 | 0.07 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr4_+_75480629 | 0.07 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr15_-_78913628 | 0.07 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr8_-_94928861 | 0.07 |
ENST00000607097.1
|
MIR378D2
|
microRNA 378d-2 |
| chrX_-_80457385 | 0.07 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr5_+_96212185 | 0.07 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr20_-_56284816 | 0.07 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr17_-_6917755 | 0.07 |
ENST00000593646.1
|
AC040977.1
|
Uncharacterized protein |
| chr6_+_150285135 | 0.07 |
ENST00000229708.3
|
ULBP1
|
UL16 binding protein 1 |
| chr20_+_46130619 | 0.07 |
ENST00000372004.3
|
NCOA3
|
nuclear receptor coactivator 3 |
| chr6_-_31648150 | 0.07 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr6_+_1312675 | 0.07 |
ENST00000296839.2
|
FOXQ1
|
forkhead box Q1 |
| chr11_-_114271139 | 0.07 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr11_+_18287801 | 0.07 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_-_60010556 | 0.07 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr1_-_153521714 | 0.06 |
ENST00000368713.3
|
S100A3
|
S100 calcium binding protein A3 |
| chr11_+_73661364 | 0.06 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr10_-_4720301 | 0.06 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr3_+_171758344 | 0.06 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr21_-_46340807 | 0.06 |
ENST00000397846.3
ENST00000524251.1 ENST00000522688.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr19_-_9811347 | 0.06 |
ENST00000585964.1
ENST00000457674.2 ENST00000590544.1 |
ZNF812
|
zinc finger protein 812 |
| chr12_-_8693469 | 0.06 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr1_-_246357029 | 0.06 |
ENST00000391836.2
|
SMYD3
|
SET and MYND domain containing 3 |
| chr11_-_6341724 | 0.06 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr5_+_1801503 | 0.06 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr5_-_39219641 | 0.06 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr19_+_51152702 | 0.06 |
ENST00000425202.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr3_-_139195350 | 0.06 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr8_+_1919481 | 0.06 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr4_-_75024085 | 0.06 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr17_-_55911970 | 0.06 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr11_+_74811578 | 0.06 |
ENST00000531713.1
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr6_-_132834184 | 0.06 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr5_+_71616188 | 0.06 |
ENST00000380639.5
ENST00000543322.1 ENST00000503868.1 ENST00000510676.2 ENST00000536805.1 |
PTCD2
|
pentatricopeptide repeat domain 2 |
| chr17_+_72199721 | 0.06 |
ENST00000439590.2
ENST00000311111.6 ENST00000584577.1 ENST00000534490.1 ENST00000528433.2 ENST00000533498.1 |
RPL38
|
ribosomal protein L38 |
| chr11_-_70963538 | 0.06 |
ENST00000413503.1
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr11_-_77734260 | 0.06 |
ENST00000353172.5
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr5_+_135394840 | 0.06 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr3_+_107602030 | 0.06 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr15_+_41099919 | 0.06 |
ENST00000561617.1
|
ZFYVE19
|
zinc finger, FYVE domain containing 19 |
| chr12_+_19358192 | 0.06 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_-_125002827 | 0.06 |
ENST00000420698.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr19_-_4182530 | 0.06 |
ENST00000601571.1
ENST00000601488.1 ENST00000305232.6 ENST00000381935.3 ENST00000337491.2 |
SIRT6
|
sirtuin 6 |
| chr15_+_45879534 | 0.06 |
ENST00000564080.1
ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4
BLOC1S6
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr6_-_109776901 | 0.06 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr6_-_32811771 | 0.06 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr4_-_7873713 | 0.06 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr11_+_107461948 | 0.06 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr12_+_1099675 | 0.06 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr4_+_74347400 | 0.06 |
ENST00000226355.3
|
AFM
|
afamin |
| chr20_+_62327996 | 0.06 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chrX_+_71401526 | 0.06 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr1_-_153521597 | 0.06 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr6_-_30043539 | 0.06 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr19_+_45458503 | 0.06 |
ENST00000337392.5
ENST00000591304.1 |
CLPTM1
|
cleft lip and palate associated transmembrane protein 1 |
| chr19_+_46002868 | 0.06 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr19_+_11670245 | 0.06 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr19_+_535835 | 0.06 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr4_-_89444883 | 0.06 |
ENST00000273968.4
|
PYURF
|
PIGY upstream reading frame |
| chr11_+_65082289 | 0.06 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr1_+_201159914 | 0.05 |
ENST00000335211.4
ENST00000451870.2 ENST00000295591.8 |
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr12_-_8693539 | 0.05 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr6_-_49712147 | 0.05 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr17_+_79071365 | 0.05 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr22_+_45725524 | 0.05 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.4 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.5 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.4 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) histone H3-K9 deacetylation(GO:1990619) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:1904180 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.0 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.0 | GO:1901074 | regulation of engulfment of apoptotic cell(GO:1901074) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.0 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.0 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.0 | 0.0 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.1 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.0 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.0 | 0.0 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.7 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0036328 | VEGF-C-activated receptor activity(GO:0036328) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |