Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
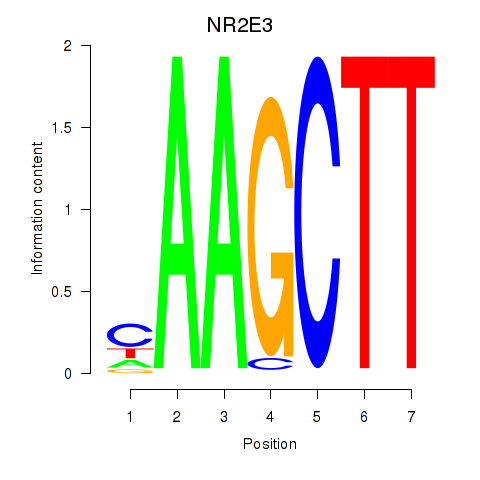
Results for NR2E3
Z-value: 1.39
Transcription factors associated with NR2E3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E3
|
ENSG00000031544.10 | nuclear receptor subfamily 2 group E member 3 |
Activity profile of NR2E3 motif
Sorted Z-values of NR2E3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_174089904 | 1.71 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr3_-_195310802 | 1.30 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr4_+_174089951 | 1.16 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr5_+_138210919 | 1.12 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr10_-_101825151 | 0.89 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr10_-_93392811 | 0.83 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chrY_+_15418467 | 0.82 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr11_+_65266507 | 0.71 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr2_-_220252068 | 0.66 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr22_-_36013368 | 0.63 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr4_-_88312301 | 0.63 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr3_+_160394940 | 0.62 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr4_-_6711558 | 0.62 |
ENST00000320848.6
|
MRFAP1L1
|
Morf4 family associated protein 1-like 1 |
| chr14_+_71165292 | 0.62 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr4_+_90033968 | 0.60 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr7_-_124569991 | 0.60 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr15_+_65914260 | 0.59 |
ENST00000261892.6
ENST00000339868.6 |
SLC24A1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr5_-_95018660 | 0.58 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr1_+_61547894 | 0.57 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chrX_-_106959631 | 0.57 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr14_+_73525265 | 0.56 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr17_+_57233087 | 0.56 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr18_-_33047039 | 0.54 |
ENST00000591141.1
ENST00000586741.1 |
RP11-322E11.5
|
RP11-322E11.5 |
| chr12_+_41221794 | 0.54 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr12_-_76477707 | 0.53 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr2_-_220252530 | 0.52 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr12_+_49740700 | 0.52 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr12_+_56477093 | 0.51 |
ENST00000549672.1
ENST00000415288.2 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_57232525 | 0.50 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_96079240 | 0.50 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr20_+_48909240 | 0.47 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr2_-_220252603 | 0.46 |
ENST00000322176.7
ENST00000273075.4 |
DNPEP
|
aspartyl aminopeptidase |
| chr2_-_197664366 | 0.46 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr1_-_179834311 | 0.46 |
ENST00000553856.1
|
IFRG15
|
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr2_+_33359687 | 0.45 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_57232690 | 0.45 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr3_+_181429704 | 0.45 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr3_+_186501336 | 0.45 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr17_-_57232596 | 0.43 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr6_-_108278456 | 0.43 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr3_+_164924716 | 0.43 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr4_-_110723335 | 0.43 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr12_+_79258444 | 0.42 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr5_+_180682720 | 0.41 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr2_+_33359646 | 0.41 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr14_-_92247032 | 0.40 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr4_+_71859156 | 0.39 |
ENST00000286648.5
ENST00000504730.1 ENST00000504952.1 |
DCK
|
deoxycytidine kinase |
| chr11_-_102962874 | 0.39 |
ENST00000531543.1
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
| chr16_+_53483983 | 0.38 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr2_+_24150180 | 0.38 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr6_-_101329191 | 0.38 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr19_-_9546177 | 0.38 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr9_+_74764340 | 0.37 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr17_-_46692457 | 0.37 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr12_-_57146095 | 0.37 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr1_+_28099683 | 0.37 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr11_-_27384737 | 0.36 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr19_+_47421933 | 0.36 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chrX_-_39956656 | 0.36 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr12_+_10658489 | 0.36 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr21_+_40752170 | 0.35 |
ENST00000333781.5
ENST00000541890.1 |
WRB
|
tryptophan rich basic protein |
| chr11_+_115498761 | 0.35 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr12_+_79258547 | 0.35 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr19_-_9546227 | 0.35 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chrX_-_48858630 | 0.34 |
ENST00000376425.3
ENST00000376444.3 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr17_+_68100989 | 0.34 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_+_74405501 | 0.33 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr21_+_17792672 | 0.33 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_-_95449155 | 0.33 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr18_+_19321281 | 0.33 |
ENST00000261537.6
|
MIB1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chrX_+_37639264 | 0.33 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr8_-_83589388 | 0.33 |
ENST00000522776.1
|
RP11-653B10.1
|
RP11-653B10.1 |
| chr12_-_110906027 | 0.32 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr4_-_110723194 | 0.32 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chrX_+_102883887 | 0.32 |
ENST00000372625.3
ENST00000372624.3 |
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr14_+_31494841 | 0.32 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr16_+_31404624 | 0.32 |
ENST00000389202.2
|
ITGAD
|
integrin, alpha D |
| chr20_-_17539456 | 0.32 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr1_+_156084461 | 0.31 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr4_-_110723134 | 0.31 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr3_-_165555200 | 0.31 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr7_-_130080818 | 0.31 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr15_+_23255242 | 0.31 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr12_-_66317967 | 0.31 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr7_-_84121858 | 0.31 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr4_-_25865159 | 0.31 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chrX_-_48858667 | 0.30 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr4_-_152147579 | 0.30 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr8_+_74903580 | 0.30 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_-_120400960 | 0.30 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr9_+_33795533 | 0.30 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr15_-_77197620 | 0.29 |
ENST00000565970.1
ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER
|
S-phase cyclin A-associated protein in the ER |
| chr4_-_23891693 | 0.29 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr18_-_28622699 | 0.29 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr11_+_20385231 | 0.29 |
ENST00000530266.1
ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr3_+_99536663 | 0.29 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr3_-_197300194 | 0.29 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr11_-_73471655 | 0.29 |
ENST00000400470.2
|
RAB6A
|
RAB6A, member RAS oncogene family |
| chr20_-_40247002 | 0.29 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr13_-_45010939 | 0.28 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr12_+_510795 | 0.28 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr17_-_27405875 | 0.28 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr18_+_42260861 | 0.28 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr18_-_28622774 | 0.27 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr5_+_75904950 | 0.27 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr14_+_73525229 | 0.27 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr3_+_9850199 | 0.27 |
ENST00000452597.1
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr12_-_51664058 | 0.27 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr22_-_37172111 | 0.27 |
ENST00000417951.2
ENST00000430701.1 ENST00000433985.2 |
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr3_-_186262166 | 0.27 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr20_-_14318248 | 0.27 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr19_+_41509851 | 0.27 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr3_-_15374033 | 0.27 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr19_+_30433372 | 0.27 |
ENST00000312051.6
|
URI1
|
URI1, prefoldin-like chaperone |
| chr14_+_24407940 | 0.26 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr12_-_76462713 | 0.26 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr7_-_155604967 | 0.26 |
ENST00000297261.2
|
SHH
|
sonic hedgehog |
| chr19_-_52307357 | 0.26 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr5_+_41925325 | 0.26 |
ENST00000296812.2
ENST00000281623.3 ENST00000509134.1 |
FBXO4
|
F-box protein 4 |
| chr12_+_96588368 | 0.26 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_149858461 | 0.26 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr1_+_84630053 | 0.26 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chrX_+_123095155 | 0.26 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr17_+_38219063 | 0.26 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chr15_+_75074410 | 0.26 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr16_+_23690138 | 0.25 |
ENST00000300093.4
|
PLK1
|
polo-like kinase 1 |
| chr7_-_103848405 | 0.25 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr16_+_2016821 | 0.25 |
ENST00000569210.2
ENST00000569714.1 |
RNF151
|
ring finger protein 151 |
| chr4_+_100737954 | 0.25 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr8_-_67579418 | 0.24 |
ENST00000310421.4
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr11_-_67141090 | 0.24 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr7_+_92158083 | 0.24 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr18_+_55712915 | 0.24 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_-_94759595 | 0.24 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_+_510742 | 0.24 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr15_+_45879964 | 0.24 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr8_+_55466915 | 0.24 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr3_-_64211112 | 0.24 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr12_+_53443963 | 0.24 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr14_-_39639523 | 0.23 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr8_+_101170563 | 0.23 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr19_-_42758040 | 0.23 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr9_+_139847347 | 0.23 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr1_-_184723942 | 0.23 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr22_-_41864662 | 0.23 |
ENST00000216252.3
|
PHF5A
|
PHD finger protein 5A |
| chr2_-_216257849 | 0.23 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr11_+_20385327 | 0.23 |
ENST00000451739.2
ENST00000532505.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr12_-_81331460 | 0.22 |
ENST00000549417.1
|
LIN7A
|
lin-7 homolog A (C. elegans) |
| chr17_+_68101117 | 0.22 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr22_-_37172191 | 0.22 |
ENST00000340630.5
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr3_-_46000064 | 0.22 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr2_+_160590469 | 0.22 |
ENST00000409591.1
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7, E3 ubiquitin protein ligase |
| chr9_-_26947453 | 0.22 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr10_-_103603523 | 0.22 |
ENST00000370046.1
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_-_124456598 | 0.22 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr12_+_49717081 | 0.22 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr4_-_156298087 | 0.22 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr7_-_111202511 | 0.21 |
ENST00000452895.1
ENST00000452753.1 ENST00000331762.3 |
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr2_+_181845763 | 0.21 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr21_+_40752378 | 0.21 |
ENST00000398753.1
ENST00000442773.1 |
WRB
|
tryptophan rich basic protein |
| chr14_+_56127960 | 0.21 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_-_91296737 | 0.21 |
ENST00000369332.3
ENST00000369329.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr14_+_90422239 | 0.21 |
ENST00000393452.3
ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1
|
tyrosyl-DNA phosphodiesterase 1 |
| chrX_-_107682702 | 0.21 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr11_-_6426635 | 0.21 |
ENST00000608645.1
ENST00000608394.1 ENST00000529519.1 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_-_20033052 | 0.21 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr3_+_37035263 | 0.21 |
ENST00000458205.2
ENST00000539477.1 |
MLH1
|
mutL homolog 1 |
| chr22_+_41865109 | 0.21 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr12_+_58138664 | 0.21 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr13_-_48575376 | 0.21 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr11_-_914774 | 0.20 |
ENST00000528154.1
ENST00000525840.1 |
CHID1
|
chitinase domain containing 1 |
| chr6_+_57182400 | 0.20 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr2_-_225434538 | 0.20 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr6_-_25874440 | 0.20 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chrX_+_37639302 | 0.20 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr14_-_54908043 | 0.20 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr6_+_42984723 | 0.20 |
ENST00000332245.8
|
KLHDC3
|
kelch domain containing 3 |
| chr14_-_94759361 | 0.20 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_94759408 | 0.20 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr14_-_37641618 | 0.20 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr3_-_97690931 | 0.20 |
ENST00000360258.4
|
MINA
|
MYC induced nuclear antigen |
| chr16_+_32264645 | 0.20 |
ENST00000569631.1
ENST00000354614.3 |
TP53TG3D
|
TP53 target 3D |
| chr1_+_43282782 | 0.20 |
ENST00000372517.2
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr2_-_175712270 | 0.20 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr1_+_87797351 | 0.20 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr12_-_15815626 | 0.19 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr11_+_20385666 | 0.19 |
ENST00000532081.1
ENST00000531058.1 |
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa |
| chr9_-_90589402 | 0.19 |
ENST00000375871.4
ENST00000605159.1 ENST00000336654.5 |
CDK20
|
cyclin-dependent kinase 20 |
| chr14_+_31494672 | 0.19 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr7_+_106809406 | 0.19 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr2_+_108905095 | 0.19 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr14_+_73525144 | 0.19 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr17_-_49124230 | 0.19 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr1_-_43282906 | 0.19 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr15_-_32747835 | 0.18 |
ENST00000509311.2
ENST00000414865.2 |
GOLGA8O
|
golgin A8 family, member O |
| chr6_-_101329157 | 0.18 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr15_-_33447055 | 0.18 |
ENST00000559047.1
ENST00000561249.1 |
FMN1
|
formin 1 |
| chr3_+_186501979 | 0.18 |
ENST00000498746.1
|
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr6_-_33864684 | 0.18 |
ENST00000506222.2
ENST00000533304.1 |
LINC01016
|
long intergenic non-protein coding RNA 1016 |
| chr20_-_40247133 | 0.18 |
ENST00000373233.3
ENST00000309279.7 |
CHD6
|
chromodomain helicase DNA binding protein 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:2000097 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 0.9 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 1.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.5 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 0.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.5 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.5 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.5 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.1 | 0.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) sclerotome development(GO:0061056) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.6 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.5 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 0.2 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.6 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.6 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.3 | GO:0017055 | female courtship behavior(GO:0008050) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 | 0.5 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.2 | GO:1904640 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.5 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:1900081 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0071874 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.5 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 2.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.6 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.4 | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.3 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.2 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 1.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.6 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.4 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.5 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.2 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.3 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 1.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 2.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |