Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
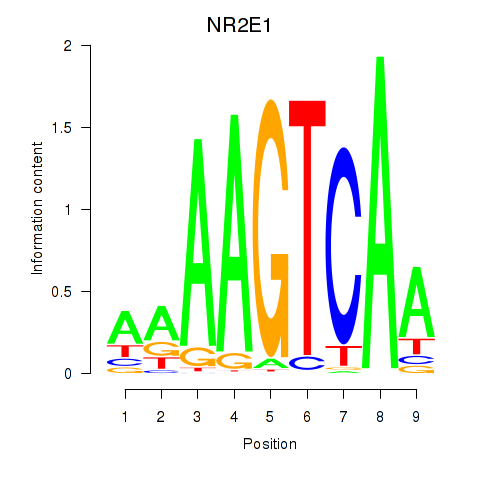
Results for NR2E1
Z-value: 0.94
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | 0.66 | 3.4e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_148939598 | 1.25 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chrY_+_15418467 | 1.11 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr16_-_66952779 | 0.94 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_-_66952742 | 0.84 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr6_+_131521120 | 0.81 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr22_-_41842781 | 0.66 |
ENST00000434408.1
|
TOB2
|
transducer of ERBB2, 2 |
| chr7_+_142829162 | 0.58 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr3_-_112356944 | 0.57 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr19_-_49567124 | 0.56 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr1_-_197036364 | 0.55 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_-_190445499 | 0.51 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr20_+_57875457 | 0.49 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chr1_+_212738676 | 0.46 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr2_-_55237484 | 0.46 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr3_-_148939835 | 0.45 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_-_90049878 | 0.42 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_-_55777586 | 0.42 |
ENST00000506836.1
|
CTC-236F12.4
|
Uncharacterized protein |
| chr12_+_40618873 | 0.42 |
ENST00000298910.7
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr12_-_51418549 | 0.41 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_+_86268065 | 0.40 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr15_+_48413211 | 0.39 |
ENST00000449382.2
|
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr14_-_92302784 | 0.39 |
ENST00000340892.5
ENST00000360594.5 |
TC2N
|
tandem C2 domains, nuclear |
| chr15_-_34502197 | 0.38 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr7_+_90012986 | 0.38 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chrX_+_47077680 | 0.38 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr9_-_99064429 | 0.38 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr12_-_111180644 | 0.37 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chrX_+_10031499 | 0.37 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr9_-_119162885 | 0.37 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr9_-_75567962 | 0.36 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr11_-_111794446 | 0.35 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_90049828 | 0.35 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr15_-_34502278 | 0.35 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr18_+_13382553 | 0.35 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr3_-_139396801 | 0.34 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr17_+_70036164 | 0.34 |
ENST00000602013.1
|
AC007461.1
|
Uncharacterized protein |
| chr22_+_25465786 | 0.34 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr7_-_130066571 | 0.33 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr17_-_46667628 | 0.33 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr4_+_52917451 | 0.32 |
ENST00000295213.4
ENST00000419395.2 |
SPATA18
|
spermatogenesis associated 18 |
| chr16_+_21608525 | 0.32 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chr2_-_45162783 | 0.32 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr17_-_46667594 | 0.32 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr11_-_63439013 | 0.32 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr15_+_23255242 | 0.31 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr11_-_63439174 | 0.31 |
ENST00000332645.4
|
ATL3
|
atlastin GTPase 3 |
| chr9_-_3469181 | 0.31 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr15_+_36887069 | 0.31 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr6_+_125540951 | 0.30 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr16_+_47495225 | 0.30 |
ENST00000299167.8
ENST00000323584.5 ENST00000563376.1 |
PHKB
|
phosphorylase kinase, beta |
| chr12_+_56624436 | 0.29 |
ENST00000266980.4
ENST00000437277.1 |
SLC39A5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr16_-_10652993 | 0.29 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr20_-_4229721 | 0.29 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr16_+_47495201 | 0.28 |
ENST00000566044.1
ENST00000455779.1 |
PHKB
|
phosphorylase kinase, beta |
| chr3_-_139396853 | 0.28 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr16_+_47496023 | 0.28 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr4_+_96012614 | 0.28 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_+_67624430 | 0.28 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr9_-_98189055 | 0.27 |
ENST00000433644.2
|
RP11-435O5.2
|
RP11-435O5.2 |
| chrX_-_106362013 | 0.27 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr12_+_79258444 | 0.27 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr20_-_45980621 | 0.27 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr15_+_48413169 | 0.26 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr21_-_46492927 | 0.26 |
ENST00000599569.1
|
AP001579.1
|
Uncharacterized protein |
| chr1_+_150229554 | 0.26 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chrX_-_128657457 | 0.26 |
ENST00000371121.3
ENST00000371123.1 ENST00000371122.4 |
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr13_-_54706954 | 0.26 |
ENST00000606706.1
ENST00000607494.1 ENST00000427299.2 ENST00000423442.2 ENST00000451744.1 |
LINC00458
|
long intergenic non-protein coding RNA 458 |
| chr5_-_42811986 | 0.26 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr12_-_68696652 | 0.26 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr2_-_111291587 | 0.26 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr1_-_108231101 | 0.25 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr16_-_15180257 | 0.24 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr10_-_115423792 | 0.24 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr6_-_10747802 | 0.24 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr14_+_24407940 | 0.24 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr14_+_31494841 | 0.24 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_+_3877412 | 0.23 |
ENST00000527651.1
|
STIM1
|
stromal interaction molecule 1 |
| chr13_+_110958124 | 0.23 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_+_179322481 | 0.23 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr12_-_31479045 | 0.23 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr4_-_153303658 | 0.23 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_+_79258547 | 0.22 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr11_-_8832182 | 0.22 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr5_+_71475449 | 0.22 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr12_-_8088773 | 0.22 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr3_+_69788576 | 0.22 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr20_+_33589048 | 0.22 |
ENST00000446156.1
ENST00000453028.1 ENST00000435272.1 ENST00000433934.2 ENST00000456649.1 |
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr8_+_104384616 | 0.22 |
ENST00000520337.1
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr14_+_39734482 | 0.21 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr2_+_207630081 | 0.20 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr4_-_157892055 | 0.20 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr9_+_2029019 | 0.20 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_58702742 | 0.19 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr14_-_71107921 | 0.19 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr7_-_5998714 | 0.19 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr12_+_72061563 | 0.19 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr3_+_141106643 | 0.19 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_108582168 | 0.19 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr5_-_142780280 | 0.19 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_+_240408560 | 0.18 |
ENST00000441342.1
ENST00000545751.1 |
FMN2
|
formin 2 |
| chr22_-_33968239 | 0.18 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr2_+_27760247 | 0.18 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr3_+_122044084 | 0.18 |
ENST00000264474.3
ENST00000479204.1 |
CSTA
|
cystatin A (stefin A) |
| chr7_-_72936608 | 0.18 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr6_+_74405501 | 0.18 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr17_+_29664830 | 0.18 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr12_+_40618764 | 0.18 |
ENST00000343742.2
|
LRRK2
|
leucine-rich repeat kinase 2 |
| chr17_+_46133307 | 0.18 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr20_-_45976816 | 0.18 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr7_-_151217166 | 0.17 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr9_-_99064386 | 0.17 |
ENST00000375262.2
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr5_+_75699149 | 0.17 |
ENST00000379730.3
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr5_-_37371163 | 0.17 |
ENST00000513532.1
|
NUP155
|
nucleoporin 155kDa |
| chr7_+_116166331 | 0.17 |
ENST00000393468.1
ENST00000393467.1 |
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr4_-_141348999 | 0.17 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr8_-_17942432 | 0.16 |
ENST00000381733.4
ENST00000314146.10 |
ASAH1
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr10_-_13350447 | 0.16 |
ENST00000429930.1
|
AL138764.1
|
Uncharacterized protein |
| chr3_+_189349162 | 0.16 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr14_+_31494672 | 0.16 |
ENST00000542754.2
ENST00000313566.6 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr17_-_29624343 | 0.16 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chrX_+_114874727 | 0.16 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr9_-_5339873 | 0.16 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr16_-_3068171 | 0.16 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr1_-_244006528 | 0.16 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr2_-_207630033 | 0.16 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr7_+_106809406 | 0.16 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr2_+_181845763 | 0.16 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr9_+_5890802 | 0.15 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr4_+_81187753 | 0.15 |
ENST00000312465.7
ENST00000456523.3 |
FGF5
|
fibroblast growth factor 5 |
| chr4_-_40631859 | 0.15 |
ENST00000295971.7
ENST00000319592.4 |
RBM47
|
RNA binding motif protein 47 |
| chr3_+_141106458 | 0.15 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr11_-_1619524 | 0.15 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr12_-_110883346 | 0.15 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr6_+_107077471 | 0.14 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr1_-_44818599 | 0.14 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr7_-_100026280 | 0.14 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr9_-_5304432 | 0.14 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr3_+_138340067 | 0.14 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_+_69928256 | 0.14 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr6_+_12290586 | 0.14 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr17_+_9479944 | 0.13 |
ENST00000396219.3
ENST00000352665.5 |
WDR16
|
WD repeat domain 16 |
| chr19_+_24009879 | 0.13 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr12_-_122018346 | 0.13 |
ENST00000377069.4
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr12_-_28124903 | 0.13 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr9_+_4662282 | 0.13 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr6_-_41039567 | 0.13 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_-_72936531 | 0.13 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr7_+_148936732 | 0.13 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chr13_+_95364963 | 0.12 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr17_-_76220740 | 0.12 |
ENST00000600484.1
|
AC087645.1
|
Uncharacterized protein |
| chr1_+_25944341 | 0.12 |
ENST00000263979.3
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr11_+_3876859 | 0.12 |
ENST00000300737.4
|
STIM1
|
stromal interaction molecule 1 |
| chr14_+_39735411 | 0.12 |
ENST00000603904.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr10_+_114709999 | 0.12 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr11_-_111781454 | 0.12 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr3_-_138048631 | 0.12 |
ENST00000484930.1
ENST00000475751.1 |
NME9
|
NME/NM23 family member 9 |
| chrX_-_138914394 | 0.12 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr8_+_91013676 | 0.12 |
ENST00000519410.1
ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr17_+_46918925 | 0.12 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr5_-_111091948 | 0.12 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr5_+_173763250 | 0.12 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr16_+_57653989 | 0.12 |
ENST00000567835.1
ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr5_-_158757895 | 0.12 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr8_+_76452097 | 0.12 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr5_+_75699040 | 0.12 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr14_+_62164340 | 0.11 |
ENST00000557538.1
ENST00000539097.1 |
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr15_-_74374891 | 0.11 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr3_-_179322416 | 0.11 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr7_-_107880508 | 0.11 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr11_+_100784231 | 0.11 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr2_-_208030295 | 0.11 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_178995021 | 0.11 |
ENST00000263733.4
|
FAM20B
|
family with sequence similarity 20, member B |
| chr15_+_31196080 | 0.11 |
ENST00000561607.1
ENST00000565466.1 |
FAN1
|
FANCD2/FANCI-associated nuclease 1 |
| chr2_-_25194476 | 0.11 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr5_+_56471592 | 0.11 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr7_+_134671234 | 0.11 |
ENST00000436302.2
ENST00000359383.3 ENST00000458078.1 ENST00000435976.2 ENST00000455283.2 |
AGBL3
|
ATP/GTP binding protein-like 3 |
| chr12_+_14518598 | 0.11 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr9_-_125240235 | 0.10 |
ENST00000259357.2
|
OR1J1
|
olfactory receptor, family 1, subfamily J, member 1 |
| chr14_+_67291158 | 0.10 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr14_-_103434869 | 0.10 |
ENST00000559043.1
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_-_169703203 | 0.10 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr3_-_50360165 | 0.10 |
ENST00000428028.1
|
HYAL2
|
hyaluronoglucosaminidase 2 |
| chr12_-_99548645 | 0.09 |
ENST00000549025.2
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_-_95846556 | 0.09 |
ENST00000426881.2
|
RP11-14O19.2
|
RP11-14O19.2 |
| chr3_-_93747425 | 0.09 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr12_+_6898674 | 0.09 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr17_-_56494713 | 0.09 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr12_+_122018697 | 0.09 |
ENST00000541574.1
|
RP13-941N14.1
|
RP13-941N14.1 |
| chr9_+_21409146 | 0.09 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr1_+_201708992 | 0.09 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr4_+_3388057 | 0.09 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr12_+_64238539 | 0.09 |
ENST00000357825.3
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1 |
| chr12_-_53343633 | 0.09 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr8_+_107593198 | 0.09 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr14_-_68157084 | 0.09 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr2_-_207629997 | 0.08 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr4_-_40632605 | 0.08 |
ENST00000514014.1
|
RBM47
|
RNA binding motif protein 47 |
| chr1_-_68698197 | 0.08 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr11_-_107436443 | 0.08 |
ENST00000429370.1
ENST00000417449.2 ENST00000428149.2 |
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr4_+_22999152 | 0.08 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.2 | 0.6 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.6 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.1 | 0.6 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.7 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.6 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.1 | 0.8 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.5 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.7 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:1903336 | intralumenal vesicle formation(GO:0070676) negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.1 | 0.4 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.1 | GO:0070101 | positive regulation of chemokine-mediated signaling pathway(GO:0070101) |
| 0.1 | 0.2 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 0.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.8 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0071880 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.6 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.5 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 1.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.7 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.6 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.0 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.5 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.2 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.3 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 0.2 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |