Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
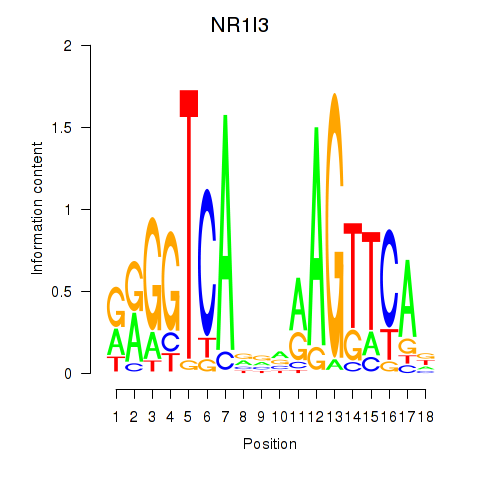
Results for NR1I3
Z-value: 0.79
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207986_161208000 | -0.34 | 6.6e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_53626974 | 0.42 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr14_+_53173890 | 0.40 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr11_+_933555 | 0.35 |
ENST00000534485.1
|
AP2A2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr20_+_5987890 | 0.34 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr12_+_100594557 | 0.32 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr1_+_145516560 | 0.29 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr1_+_145516252 | 0.29 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr11_-_71781096 | 0.29 |
ENST00000535087.1
ENST00000535838.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr9_-_35361262 | 0.26 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chr14_+_53173910 | 0.25 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr5_+_139055021 | 0.24 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr11_-_104480019 | 0.24 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr19_+_9203855 | 0.24 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr17_-_62503015 | 0.23 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr16_+_53133070 | 0.22 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_143833918 | 0.21 |
ENST00000359228.3
|
LYPD2
|
LY6/PLAUR domain containing 2 |
| chr5_-_138780159 | 0.20 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr5_+_139055055 | 0.19 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr19_-_58919815 | 0.18 |
ENST00000597980.1
|
CTD-2619J13.14
|
CTD-2619J13.14 |
| chr11_-_82611448 | 0.18 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr1_-_21625486 | 0.18 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_-_235116495 | 0.18 |
ENST00000549744.1
|
RP11-443B7.3
|
RP11-443B7.3 |
| chr1_-_33336414 | 0.17 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr1_+_82165350 | 0.17 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr18_-_23670546 | 0.16 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr18_-_19284724 | 0.15 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr18_-_19283649 | 0.15 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr12_-_118810688 | 0.15 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr15_+_96875657 | 0.15 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_33390667 | 0.15 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_+_45274154 | 0.14 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr12_+_53817633 | 0.14 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr1_-_225616515 | 0.14 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr18_-_23671139 | 0.14 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr7_-_56160625 | 0.14 |
ENST00000446428.1
ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1
|
phosphorylase kinase, gamma 1 (muscle) |
| chr2_-_228028829 | 0.13 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr16_+_58535372 | 0.13 |
ENST00000566656.1
ENST00000566618.1 |
NDRG4
|
NDRG family member 4 |
| chr13_-_114107839 | 0.12 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr2_-_209118974 | 0.12 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr9_+_134001455 | 0.12 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr5_-_39270725 | 0.12 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr13_+_60971080 | 0.12 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr6_-_127780510 | 0.12 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr6_+_96969672 | 0.11 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr1_+_154377669 | 0.11 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr2_-_86790472 | 0.11 |
ENST00000409727.1
|
CHMP3
|
charged multivesicular body protein 3 |
| chr1_+_153330322 | 0.11 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr12_-_108991812 | 0.10 |
ENST00000392806.3
ENST00000547567.1 |
TMEM119
|
transmembrane protein 119 |
| chr11_-_71753188 | 0.10 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr16_+_67280799 | 0.10 |
ENST00000566345.2
|
SLC9A5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr14_+_21525981 | 0.10 |
ENST00000308227.2
|
RNASE8
|
ribonuclease, RNase A family, 8 |
| chr19_-_49140609 | 0.10 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_+_57998400 | 0.10 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr6_+_71276596 | 0.10 |
ENST00000370474.3
|
C6orf57
|
chromosome 6 open reading frame 57 |
| chr11_-_61124776 | 0.10 |
ENST00000542361.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr17_-_53499310 | 0.10 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr19_-_7562334 | 0.08 |
ENST00000593942.1
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma |
| chr17_-_62502399 | 0.08 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr10_+_71444655 | 0.08 |
ENST00000434931.2
|
RP11-242G20.1
|
Uncharacterized protein |
| chr22_+_39493207 | 0.08 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr4_+_70894130 | 0.08 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr1_+_32930647 | 0.07 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr6_+_33422343 | 0.07 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr9_-_80646374 | 0.07 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr14_-_50506589 | 0.07 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr2_-_86790593 | 0.07 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr22_+_39493268 | 0.06 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr6_-_56258892 | 0.06 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chrX_-_153583257 | 0.06 |
ENST00000438732.1
|
FLNA
|
filamin A, alpha |
| chr17_-_53499218 | 0.06 |
ENST00000571578.1
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr5_-_176937351 | 0.05 |
ENST00000377112.4
ENST00000501403.2 ENST00000312943.6 |
DOK3
|
docking protein 3 |
| chr17_-_62502639 | 0.05 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr5_+_52856456 | 0.05 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr5_-_176936817 | 0.05 |
ENST00000502885.1
ENST00000506493.1 |
DOK3
|
docking protein 3 |
| chr4_-_82136114 | 0.05 |
ENST00000395578.1
ENST00000418486.2 |
PRKG2
|
protein kinase, cGMP-dependent, type II |
| chrX_+_118602363 | 0.04 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chrX_+_110924346 | 0.04 |
ENST00000371979.3
ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr14_-_22005062 | 0.04 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr5_+_156712372 | 0.04 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr9_-_114937676 | 0.04 |
ENST00000374270.3
|
SUSD1
|
sushi domain containing 1 |
| chr6_-_43496605 | 0.04 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr19_-_6604094 | 0.04 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr9_+_124048864 | 0.04 |
ENST00000545652.1
|
GSN
|
gelsolin |
| chr19_+_12203069 | 0.04 |
ENST00000430298.2
ENST00000339302.4 |
ZNF788
ZNF788
|
zinc finger family member 788 Zinc finger protein 788 |
| chr1_+_26036093 | 0.03 |
ENST00000374329.1
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr2_-_128642434 | 0.03 |
ENST00000393001.1
|
AMMECR1L
|
AMMECR1-like |
| chr14_-_22005018 | 0.03 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_+_63137251 | 0.03 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chr11_-_61124711 | 0.03 |
ENST00000536915.1
|
CYB561A3
|
cytochrome b561 family, member A3 |
| chr11_-_57089774 | 0.03 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chrX_+_69674943 | 0.03 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr19_-_46285646 | 0.03 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr13_-_20806440 | 0.02 |
ENST00000400066.3
ENST00000400065.3 ENST00000356192.6 |
GJB6
|
gap junction protein, beta 6, 30kDa |
| chr1_-_1690014 | 0.02 |
ENST00000400922.2
ENST00000342348.5 |
NADK
|
NAD kinase |
| chr9_-_75567962 | 0.02 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr4_+_146402346 | 0.02 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr1_-_9086404 | 0.02 |
ENST00000400906.1
|
SLC2A7
|
solute carrier family 2 (facilitated glucose transporter), member 7 |
| chr19_+_58920102 | 0.01 |
ENST00000599238.1
ENST00000322834.7 |
ZNF584
|
zinc finger protein 584 |
| chr1_-_23886285 | 0.01 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr10_+_47658234 | 0.01 |
ENST00000447511.2
ENST00000537271.1 |
ANTXRL
|
anthrax toxin receptor-like |
| chr7_-_99381798 | 0.01 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr9_-_91793675 | 0.01 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr3_-_58419537 | 0.01 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr12_-_53729525 | 0.01 |
ENST00000303846.3
|
SP7
|
Sp7 transcription factor |
| chr16_-_16317321 | 0.01 |
ENST00000205557.7
ENST00000575728.1 |
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr15_+_96876340 | 0.00 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.4 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.3 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.2 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.4 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.3 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |