Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
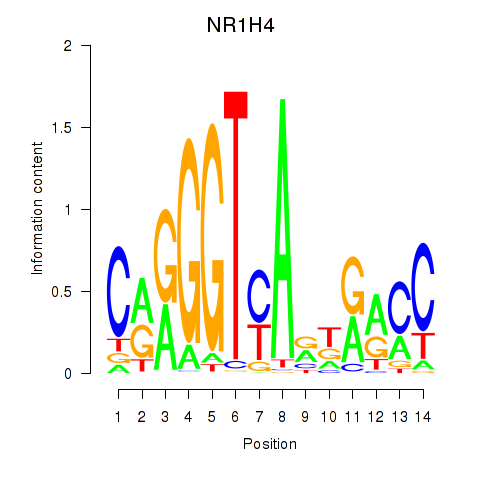
Results for NR1H4
Z-value: 0.96
Transcription factors associated with NR1H4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H4
|
ENSG00000012504.9 | nuclear receptor subfamily 1 group H member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H4 | hg19_v2_chr12_+_100867733_100867750 | -0.91 | 8.7e-02 | Click! |
Activity profile of NR1H4 motif
Sorted Z-values of NR1H4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_182545603 | 1.19 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr8_-_131028782 | 0.70 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr19_+_10197463 | 0.66 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr19_+_38880695 | 0.50 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr2_-_74374995 | 0.46 |
ENST00000295326.4
|
BOLA3
|
bolA family member 3 |
| chr6_+_107349392 | 0.41 |
ENST00000443043.1
ENST00000405204.2 ENST00000311381.5 |
C6orf203
|
chromosome 6 open reading frame 203 |
| chr11_-_6341724 | 0.41 |
ENST00000530979.1
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr14_+_57671888 | 0.40 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr1_+_1334914 | 0.39 |
ENST00000576232.1
ENST00000570344.1 |
RP4-758J18.2
|
HCG20425, isoform CRA_a; Uncharacterized protein; cDNA FLJ53815 |
| chr19_+_10196981 | 0.38 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr16_+_2014941 | 0.37 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr15_+_40532723 | 0.35 |
ENST00000558878.1
ENST00000558183.1 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr11_-_10879593 | 0.35 |
ENST00000528289.1
ENST00000432999.2 |
ZBED5
|
zinc finger, BED-type containing 5 |
| chr6_-_38607673 | 0.34 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr1_+_28206150 | 0.34 |
ENST00000456990.1
|
THEMIS2
|
thymocyte selection associated family member 2 |
| chr8_+_9046503 | 0.33 |
ENST00000512942.2
|
RP11-10A14.5
|
RP11-10A14.5 |
| chr16_+_81478775 | 0.31 |
ENST00000537098.3
|
CMIP
|
c-Maf inducing protein |
| chr19_-_44860820 | 0.31 |
ENST00000354340.4
ENST00000337401.4 ENST00000587909.1 |
ZNF112
|
zinc finger protein 112 |
| chr4_-_171011323 | 0.30 |
ENST00000509167.1
ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT
|
aminoadipate aminotransferase |
| chr11_-_64511575 | 0.29 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr11_-_118305921 | 0.28 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr12_+_57914481 | 0.28 |
ENST00000548887.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_-_71003568 | 0.28 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr17_-_73840614 | 0.27 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr2_+_192543153 | 0.26 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr11_-_10879572 | 0.26 |
ENST00000413761.2
|
ZBED5
|
zinc finger, BED-type containing 5 |
| chr5_+_131593364 | 0.25 |
ENST00000253754.3
ENST00000379018.3 |
PDLIM4
|
PDZ and LIM domain 4 |
| chr19_-_633576 | 0.25 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr5_+_140588269 | 0.24 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr3_+_122296465 | 0.24 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr1_-_36789755 | 0.24 |
ENST00000270824.1
|
EVA1B
|
eva-1 homolog B (C. elegans) |
| chr19_+_1495362 | 0.24 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr10_-_95360983 | 0.23 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr16_+_30953696 | 0.23 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr11_-_71639613 | 0.23 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr15_+_89921280 | 0.23 |
ENST00000560596.1
ENST00000558692.1 ENST00000538734.2 ENST00000559235.1 |
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr22_+_20104947 | 0.23 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr8_-_23261589 | 0.22 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chrX_-_47930980 | 0.22 |
ENST00000442455.3
ENST00000428686.1 ENST00000276054.4 |
ZNF630
|
zinc finger protein 630 |
| chr2_-_178937478 | 0.21 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chr5_+_138629337 | 0.21 |
ENST00000394805.3
ENST00000512876.1 ENST00000513678.1 |
MATR3
|
matrin 3 |
| chr3_-_28390581 | 0.21 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr3_-_57113281 | 0.21 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr12_-_68845165 | 0.21 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr19_-_11689752 | 0.20 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr1_+_162351503 | 0.20 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr14_-_21945057 | 0.20 |
ENST00000397762.1
|
RAB2B
|
RAB2B, member RAS oncogene family |
| chr1_+_14026671 | 0.20 |
ENST00000484063.2
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr2_+_192542850 | 0.19 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_-_16035195 | 0.19 |
ENST00000535752.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_+_179345173 | 0.19 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr19_-_51054299 | 0.19 |
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B |
| chr6_+_30594619 | 0.18 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr2_+_192543694 | 0.18 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_+_139847347 | 0.18 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr20_-_3644046 | 0.18 |
ENST00000290417.2
ENST00000319242.3 |
GFRA4
|
GDNF family receptor alpha 4 |
| chr5_+_132208014 | 0.18 |
ENST00000296877.2
|
LEAP2
|
liver expressed antimicrobial peptide 2 |
| chr20_+_30640004 | 0.17 |
ENST00000520553.1
ENST00000518730.1 ENST00000375852.2 |
HCK
|
hemopoietic cell kinase |
| chr9_+_74526532 | 0.17 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr4_+_26322409 | 0.17 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_67188642 | 0.17 |
ENST00000546202.1
ENST00000542876.1 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr9_-_34381511 | 0.17 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr1_+_75600567 | 0.17 |
ENST00000356261.3
|
LHX8
|
LIM homeobox 8 |
| chr2_+_192542879 | 0.16 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr20_+_30639991 | 0.16 |
ENST00000534862.1
ENST00000538448.1 ENST00000375862.2 |
HCK
|
hemopoietic cell kinase |
| chr3_+_183873098 | 0.16 |
ENST00000313143.3
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr5_-_139944196 | 0.16 |
ENST00000357560.4
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr12_-_7281469 | 0.16 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr11_-_71639446 | 0.16 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr16_+_67700673 | 0.16 |
ENST00000403458.4
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr19_-_51893782 | 0.15 |
ENST00000570516.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr17_-_3595042 | 0.15 |
ENST00000552723.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr11_-_62380199 | 0.15 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr3_-_120461378 | 0.15 |
ENST00000273375.3
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr10_+_105726862 | 0.15 |
ENST00000335753.4
ENST00000369755.3 |
SLK
|
STE20-like kinase |
| chr6_-_106773610 | 0.14 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr19_+_41117770 | 0.14 |
ENST00000601032.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_+_43318434 | 0.14 |
ENST00000587489.1
|
FMNL1
|
formin-like 1 |
| chr7_-_33140498 | 0.14 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr22_+_30163340 | 0.14 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_+_203765437 | 0.14 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr1_+_33231221 | 0.14 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr19_+_677885 | 0.13 |
ENST00000591552.2
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
| chr1_-_109584608 | 0.13 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr1_-_153585539 | 0.13 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chr15_+_83776137 | 0.13 |
ENST00000322019.9
|
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr10_-_119806085 | 0.13 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr3_+_49058444 | 0.13 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_-_42636617 | 0.13 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr4_+_123073464 | 0.13 |
ENST00000264501.4
|
KIAA1109
|
KIAA1109 |
| chr19_-_48867291 | 0.13 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr21_+_35553045 | 0.13 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr5_+_138629628 | 0.12 |
ENST00000508689.1
ENST00000514528.1 |
MATR3
|
matrin 3 |
| chr7_-_107883678 | 0.12 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr9_-_44402427 | 0.12 |
ENST00000540551.1
|
BX088651.1
|
LOC100126582 protein; Uncharacterized protein |
| chr1_-_109584716 | 0.12 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chr2_-_133104839 | 0.12 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr11_-_71639670 | 0.12 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr16_+_69166418 | 0.11 |
ENST00000314423.7
ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr19_-_51893827 | 0.11 |
ENST00000574814.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chrX_-_153141783 | 0.11 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr3_-_126236605 | 0.11 |
ENST00000290868.2
|
UROC1
|
urocanate hydratase 1 |
| chr20_-_45980621 | 0.11 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr8_-_110346614 | 0.11 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr3_-_128294929 | 0.11 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr1_-_109584768 | 0.10 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chr5_+_138629389 | 0.10 |
ENST00000504045.1
ENST00000504311.1 ENST00000502499.1 |
MATR3
|
matrin 3 |
| chrX_-_48823165 | 0.10 |
ENST00000419374.1
|
KCND1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr11_-_72070206 | 0.10 |
ENST00000544382.1
|
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chrX_-_153141434 | 0.10 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr12_-_56727676 | 0.09 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr11_+_67798114 | 0.09 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr17_-_33390667 | 0.09 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr1_-_50489547 | 0.09 |
ENST00000371836.1
ENST00000371839.1 ENST00000371838.1 |
AGBL4
|
ATP/GTP binding protein-like 4 |
| chr8_+_35649365 | 0.09 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr11_-_65429891 | 0.09 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr3_+_183873240 | 0.08 |
ENST00000431765.1
|
DVL3
|
dishevelled segment polarity protein 3 |
| chr10_-_49701686 | 0.08 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_-_74780176 | 0.08 |
ENST00000409549.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr12_+_100967533 | 0.08 |
ENST00000550295.1
|
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr2_+_58274001 | 0.08 |
ENST00000428021.1
|
VRK2
|
vaccinia related kinase 2 |
| chr14_-_21492251 | 0.08 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr19_-_35626104 | 0.08 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr17_-_73975198 | 0.08 |
ENST00000301608.4
ENST00000588176.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr3_-_160117035 | 0.08 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr12_-_56727487 | 0.08 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr5_-_16509101 | 0.08 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr12_-_8088773 | 0.08 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr3_+_111260856 | 0.08 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr19_+_1071203 | 0.08 |
ENST00000543365.1
|
HMHA1
|
histocompatibility (minor) HA-1 |
| chr4_+_25915822 | 0.08 |
ENST00000506197.2
|
SMIM20
|
small integral membrane protein 20 |
| chr1_-_16939976 | 0.07 |
ENST00000430580.2
|
NBPF1
|
neuroblastoma breakpoint family, member 1 |
| chr18_+_3448455 | 0.07 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr5_+_67576109 | 0.07 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr14_-_54908043 | 0.07 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr4_-_69817481 | 0.07 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_+_67576062 | 0.07 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr9_+_74526384 | 0.07 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr6_+_108616054 | 0.07 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr3_+_111260954 | 0.07 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr17_-_7145475 | 0.07 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr17_-_46035187 | 0.07 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr2_+_108994633 | 0.07 |
ENST00000409309.3
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr11_-_4414880 | 0.07 |
ENST00000254436.7
ENST00000543625.1 |
TRIM21
|
tripartite motif containing 21 |
| chr6_-_106773491 | 0.06 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr3_+_52489503 | 0.06 |
ENST00000345716.4
|
NISCH
|
nischarin |
| chr14_-_65769392 | 0.06 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr1_-_33642151 | 0.06 |
ENST00000543586.1
|
TRIM62
|
tripartite motif containing 62 |
| chrX_+_7137475 | 0.06 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr6_-_106773291 | 0.06 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr18_-_67624160 | 0.06 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr1_-_1334685 | 0.06 |
ENST00000400809.3
ENST00000408918.4 |
CCNL2
|
cyclin L2 |
| chr16_+_23847339 | 0.06 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr16_-_30537839 | 0.06 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chrX_+_152224766 | 0.06 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr11_+_7041677 | 0.06 |
ENST00000299481.4
|
NLRP14
|
NLR family, pyrin domain containing 14 |
| chr3_-_48632593 | 0.06 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr17_-_4852332 | 0.06 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr17_+_48351785 | 0.06 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr8_-_131028869 | 0.06 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr18_-_3880051 | 0.05 |
ENST00000584874.1
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr7_+_2671568 | 0.05 |
ENST00000258796.7
|
TTYH3
|
tweety family member 3 |
| chrX_+_103031758 | 0.05 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr7_-_131241361 | 0.05 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr2_-_219433014 | 0.05 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr12_+_120875910 | 0.05 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr19_+_11546153 | 0.05 |
ENST00000591946.1
ENST00000252455.2 ENST00000412601.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr5_-_137878887 | 0.05 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr8_-_116504448 | 0.05 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_-_35104185 | 0.05 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr19_-_48867171 | 0.05 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr7_+_2671663 | 0.05 |
ENST00000407643.1
|
TTYH3
|
tweety family member 3 |
| chr3_-_126236588 | 0.05 |
ENST00000383579.3
|
UROC1
|
urocanate hydratase 1 |
| chr13_+_27844464 | 0.05 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr20_-_45981138 | 0.05 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr11_-_33795893 | 0.04 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr3_+_186560462 | 0.04 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr9_+_127023704 | 0.04 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr3_-_28390415 | 0.04 |
ENST00000414162.1
ENST00000420543.2 |
AZI2
|
5-azacytidine induced 2 |
| chr1_-_149908217 | 0.04 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr12_-_8088871 | 0.04 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr9_-_34381536 | 0.04 |
ENST00000379126.3
ENST00000379127.1 ENST00000379133.3 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr19_+_11546440 | 0.04 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr4_+_152330390 | 0.04 |
ENST00000503146.1
ENST00000435205.1 |
FAM160A1
|
family with sequence similarity 160, member A1 |
| chr19_+_11546093 | 0.04 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr5_+_138629417 | 0.04 |
ENST00000510056.1
ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3
|
matrin 3 |
| chr6_+_127587755 | 0.04 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr15_+_65337708 | 0.03 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr3_-_15540055 | 0.03 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr6_+_4087664 | 0.03 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr3_+_54157480 | 0.03 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr3_+_52489606 | 0.03 |
ENST00000488380.1
ENST00000420808.2 |
NISCH
|
nischarin |
| chr6_-_49681235 | 0.03 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chrX_-_153141302 | 0.03 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr3_+_186560476 | 0.03 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr17_-_43568062 | 0.03 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr16_+_89238149 | 0.03 |
ENST00000289746.2
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chrX_+_103031421 | 0.03 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr8_+_110346546 | 0.03 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr11_+_67250490 | 0.03 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.7 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.2 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.3 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0002729 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.3 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |