Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
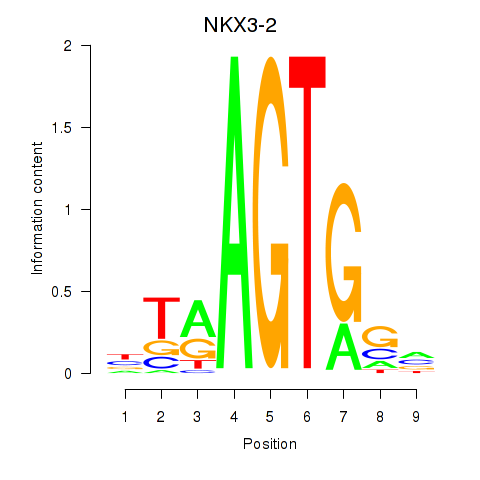
Results for NKX3-2
Z-value: 0.93
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | 0.34 | 6.6e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_42160486 | 0.48 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr12_-_113658892 | 0.35 |
ENST00000299732.2
ENST00000416617.2 |
IQCD
|
IQ motif containing D |
| chr19_+_54609230 | 0.35 |
ENST00000420296.1
|
NDUFA3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr12_-_54978086 | 0.33 |
ENST00000553113.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr12_+_34175398 | 0.32 |
ENST00000538927.1
|
ALG10
|
ALG10, alpha-1,2-glucosyltransferase |
| chrX_+_48660565 | 0.32 |
ENST00000413163.2
ENST00000441703.1 ENST00000426196.1 |
HDAC6
|
histone deacetylase 6 |
| chr6_+_32821924 | 0.29 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_145780725 | 0.28 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chrX_+_48660107 | 0.27 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr19_+_18544045 | 0.27 |
ENST00000599699.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr19_+_7694623 | 0.26 |
ENST00000594797.1
ENST00000456958.3 ENST00000601406.1 |
PET100
|
PET100 homolog (S. cerevisiae) |
| chr1_+_119957554 | 0.26 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_-_25451065 | 0.25 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr9_-_131940526 | 0.25 |
ENST00000372491.2
|
IER5L
|
immediate early response 5-like |
| chr22_+_38453207 | 0.22 |
ENST00000404072.3
ENST00000424694.1 |
PICK1
|
protein interacting with PRKCA 1 |
| chr2_-_188312971 | 0.21 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr11_+_59705928 | 0.21 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr3_+_48481658 | 0.20 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chrX_+_48660287 | 0.20 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr6_+_26156551 | 0.20 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr9_+_35906176 | 0.20 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr10_-_131762105 | 0.20 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr3_-_146262293 | 0.20 |
ENST00000448205.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr5_-_175965008 | 0.19 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr12_-_21928515 | 0.19 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr15_-_40401062 | 0.19 |
ENST00000354670.4
ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF
|
Bcl2 modifying factor |
| chr1_+_26606608 | 0.19 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_-_10444188 | 0.19 |
ENST00000293677.6
|
RAVER1
|
ribonucleoprotein, PTB-binding 1 |
| chr9_-_95244781 | 0.18 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr6_+_80129989 | 0.18 |
ENST00000429444.1
|
RP1-232L24.3
|
RP1-232L24.3 |
| chr8_+_67624653 | 0.18 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr1_-_1590418 | 0.17 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr12_+_4382917 | 0.17 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr19_-_5680231 | 0.17 |
ENST00000587950.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr19_-_38747172 | 0.17 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr7_-_102119342 | 0.16 |
ENST00000393794.3
ENST00000292614.5 |
POLR2J
|
polymerase (RNA) II (DNA directed) polypeptide J, 13.3kDa |
| chr6_+_37012607 | 0.16 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr11_+_76813886 | 0.16 |
ENST00000529803.1
|
OMP
|
olfactory marker protein |
| chr2_+_220379052 | 0.15 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr5_+_179220979 | 0.15 |
ENST00000292596.10
ENST00000401985.3 |
LTC4S
|
leukotriene C4 synthase |
| chr1_+_10509971 | 0.15 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr19_+_40477062 | 0.15 |
ENST00000455878.2
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr20_+_1115821 | 0.15 |
ENST00000435720.1
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr2_+_177015950 | 0.14 |
ENST00000306324.3
|
HOXD4
|
homeobox D4 |
| chr22_-_21356375 | 0.14 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr11_+_89867681 | 0.14 |
ENST00000534061.1
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr22_+_38453378 | 0.14 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr7_+_30174668 | 0.14 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_112218378 | 0.14 |
ENST00000334529.5
|
BTLA
|
B and T lymphocyte associated |
| chr9_+_134065506 | 0.14 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr12_+_109569155 | 0.14 |
ENST00000539864.1
|
ACACB
|
acetyl-CoA carboxylase beta |
| chr20_+_2276639 | 0.13 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr22_-_31328881 | 0.13 |
ENST00000445980.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr2_-_25016251 | 0.13 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr7_+_73106926 | 0.13 |
ENST00000453316.1
|
WBSCR22
|
Williams Beuren syndrome chromosome region 22 |
| chr2_+_25016282 | 0.13 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr14_-_67859422 | 0.13 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr5_-_137514333 | 0.13 |
ENST00000411594.2
ENST00000430331.1 |
BRD8
|
bromodomain containing 8 |
| chr6_+_42928485 | 0.13 |
ENST00000372808.3
|
GNMT
|
glycine N-methyltransferase |
| chr19_-_5680202 | 0.12 |
ENST00000590389.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr3_+_169629354 | 0.12 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr1_-_202858227 | 0.12 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr1_-_226065330 | 0.12 |
ENST00000436966.1
|
TMEM63A
|
transmembrane protein 63A |
| chr1_-_155145721 | 0.12 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr2_-_38978492 | 0.12 |
ENST00000409276.1
ENST00000446327.2 ENST00000313117.6 |
SRSF7
|
serine/arginine-rich splicing factor 7 |
| chr3_-_146262428 | 0.12 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_+_156756667 | 0.12 |
ENST00000526188.1
ENST00000454659.1 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr18_-_3220106 | 0.12 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr3_-_50541028 | 0.12 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr11_+_64879317 | 0.12 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr22_+_31489344 | 0.12 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr8_-_37707356 | 0.12 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr7_+_110731062 | 0.12 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chrX_+_51928002 | 0.11 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr19_+_36545833 | 0.11 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr2_+_149402009 | 0.11 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr10_+_35415851 | 0.11 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr6_-_32821599 | 0.11 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr18_+_21464737 | 0.11 |
ENST00000586751.1
|
LAMA3
|
laminin, alpha 3 |
| chrX_-_15332665 | 0.11 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr12_+_6309963 | 0.11 |
ENST00000382515.2
|
CD9
|
CD9 molecule |
| chr20_+_2821366 | 0.11 |
ENST00000453689.1
ENST00000417508.1 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr16_+_73420942 | 0.11 |
ENST00000554640.1
ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1
|
RP11-140I24.1 |
| chr9_+_34458851 | 0.11 |
ENST00000545019.1
|
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr12_-_68845165 | 0.11 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr17_-_74137374 | 0.11 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr17_+_7482785 | 0.11 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr3_-_116163830 | 0.11 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr11_-_119993979 | 0.11 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr19_+_48867652 | 0.11 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4 |
| chr9_+_116263778 | 0.11 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr17_-_7145475 | 0.11 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr15_-_90777277 | 0.11 |
ENST00000328649.6
|
CIB1
|
calcium and integrin binding 1 (calmyrin) |
| chr19_+_39109735 | 0.11 |
ENST00000593149.1
ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K
|
eukaryotic translation initiation factor 3, subunit K |
| chr15_+_81475047 | 0.11 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr2_+_62132781 | 0.11 |
ENST00000311832.5
|
COMMD1
|
copper metabolism (Murr1) domain containing 1 |
| chr1_-_217262969 | 0.11 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr1_-_156710916 | 0.11 |
ENST00000368211.4
|
MRPL24
|
mitochondrial ribosomal protein L24 |
| chr16_-_28222797 | 0.11 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr3_+_48507210 | 0.11 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr14_+_78227105 | 0.10 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr12_-_120765565 | 0.10 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr1_+_162351503 | 0.10 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr19_-_6720686 | 0.10 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr8_-_49833978 | 0.10 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr8_+_86376081 | 0.10 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr6_+_24357131 | 0.10 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr9_-_80645520 | 0.10 |
ENST00000411677.1
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr14_-_24610779 | 0.10 |
ENST00000560403.1
ENST00000419198.2 ENST00000216799.4 |
EMC9
|
ER membrane protein complex subunit 9 |
| chr20_+_2821340 | 0.10 |
ENST00000380445.3
ENST00000380469.3 |
VPS16
|
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr11_-_26593649 | 0.10 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr19_-_18548921 | 0.10 |
ENST00000545187.1
ENST00000578352.1 |
ISYNA1
|
inositol-3-phosphate synthase 1 |
| chr10_-_17171817 | 0.09 |
ENST00000377833.4
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr12_+_53693466 | 0.09 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr4_-_168155700 | 0.09 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr17_+_30593195 | 0.09 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr11_+_66045634 | 0.09 |
ENST00000528852.1
ENST00000311445.6 |
CNIH2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr10_-_46168156 | 0.09 |
ENST00000374371.2
ENST00000335258.7 |
ZFAND4
|
zinc finger, AN1-type domain 4 |
| chr19_+_14672755 | 0.09 |
ENST00000594545.1
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr3_-_100551141 | 0.09 |
ENST00000478235.1
ENST00000471901.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr2_-_208489707 | 0.09 |
ENST00000448007.2
ENST00000432416.1 ENST00000411432.1 |
METTL21A
|
methyltransferase like 21A |
| chr11_+_2421718 | 0.09 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr11_-_107590383 | 0.09 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr7_-_8276508 | 0.09 |
ENST00000401396.1
ENST00000317367.5 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr5_-_137514288 | 0.09 |
ENST00000454473.1
ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8
|
bromodomain containing 8 |
| chr1_-_78444738 | 0.09 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr7_-_22862406 | 0.09 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr2_+_74229812 | 0.09 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr21_-_33985127 | 0.08 |
ENST00000290155.3
|
C21orf59
|
chromosome 21 open reading frame 59 |
| chr6_-_27279949 | 0.08 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr4_-_168155730 | 0.08 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr9_+_34458771 | 0.08 |
ENST00000437363.1
ENST00000242317.4 |
DNAI1
|
dynein, axonemal, intermediate chain 1 |
| chr12_-_113658826 | 0.08 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr2_-_85625857 | 0.08 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr6_-_30080876 | 0.08 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chrX_-_49965663 | 0.08 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr2_-_70780770 | 0.08 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr10_+_105005644 | 0.08 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr8_-_29592736 | 0.08 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr4_-_4228616 | 0.08 |
ENST00000296358.4
|
OTOP1
|
otopetrin 1 |
| chr21_-_43816052 | 0.08 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr17_-_42295870 | 0.08 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr10_+_47746929 | 0.08 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr10_-_47173994 | 0.08 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr21_+_33671160 | 0.08 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_+_177545563 | 0.08 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr11_+_35222629 | 0.08 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr6_-_32784687 | 0.08 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr20_+_53092232 | 0.08 |
ENST00000395939.1
|
DOK5
|
docking protein 5 |
| chr2_-_27294500 | 0.08 |
ENST00000447619.1
ENST00000429985.1 ENST00000456793.1 |
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
| chr11_-_128737163 | 0.08 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr14_+_64565442 | 0.08 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_+_57984943 | 0.08 |
ENST00000422156.3
|
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr7_+_90339169 | 0.08 |
ENST00000436577.2
|
CDK14
|
cyclin-dependent kinase 14 |
| chrX_-_103087136 | 0.08 |
ENST00000243298.2
|
RAB9B
|
RAB9B, member RAS oncogene family |
| chr1_-_31769595 | 0.08 |
ENST00000263694.4
|
SNRNP40
|
small nuclear ribonucleoprotein 40kDa (U5) |
| chr15_+_75640068 | 0.07 |
ENST00000565051.1
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr19_-_17185848 | 0.07 |
ENST00000593360.1
|
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr12_-_62586543 | 0.07 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr11_+_67250490 | 0.07 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr12_-_95397442 | 0.07 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr20_+_53092123 | 0.07 |
ENST00000262593.5
|
DOK5
|
docking protein 5 |
| chr1_+_149230680 | 0.07 |
ENST00000443018.1
|
RP11-403I13.5
|
RP11-403I13.5 |
| chr12_-_57037284 | 0.07 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr8_-_16859690 | 0.07 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr14_-_50778931 | 0.07 |
ENST00000555423.1
ENST00000421284.3 |
L2HGDH
|
L-2-hydroxyglutarate dehydrogenase |
| chr12_+_50794891 | 0.07 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr18_-_13915530 | 0.07 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr8_-_49834299 | 0.07 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr7_+_129007964 | 0.07 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_+_120629653 | 0.07 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr3_-_112218205 | 0.07 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated |
| chr3_+_111697843 | 0.07 |
ENST00000534857.1
ENST00000273359.3 ENST00000494817.1 |
ABHD10
|
abhydrolase domain containing 10 |
| chr18_+_59000815 | 0.07 |
ENST00000262717.4
|
CDH20
|
cadherin 20, type 2 |
| chr7_-_79082867 | 0.07 |
ENST00000419488.1
ENST00000354212.4 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr20_+_54987168 | 0.07 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr1_+_164528437 | 0.07 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr12_+_57984965 | 0.07 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr14_+_23016437 | 0.07 |
ENST00000478163.3
|
TRAC
|
T cell receptor alpha constant |
| chr10_-_95360983 | 0.07 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr10_-_75571341 | 0.07 |
ENST00000309979.6
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr20_-_62493217 | 0.07 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr6_-_33663474 | 0.07 |
ENST00000594414.1
|
SBP1
|
SBP1; Uncharacterized protein |
| chr11_-_64511789 | 0.07 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_-_73150629 | 0.07 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr5_-_115872142 | 0.07 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr17_-_46507537 | 0.07 |
ENST00000336915.6
|
SKAP1
|
src kinase associated phosphoprotein 1 |
| chr16_-_20566616 | 0.07 |
ENST00000569163.1
|
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr2_+_145780767 | 0.07 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr7_-_102213030 | 0.07 |
ENST00000511313.1
ENST00000513438.1 ENST00000513506.1 |
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr11_-_59436453 | 0.06 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr19_+_843314 | 0.06 |
ENST00000544537.2
|
PRTN3
|
proteinase 3 |
| chr10_-_75571566 | 0.06 |
ENST00000299641.4
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr17_-_60883993 | 0.06 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr8_-_123706338 | 0.06 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr10_+_35416090 | 0.06 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr5_+_112074029 | 0.06 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr5_-_59481406 | 0.06 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr4_-_48018680 | 0.06 |
ENST00000513178.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr17_+_18855470 | 0.06 |
ENST00000395647.2
ENST00000395642.1 ENST00000417251.2 ENST00000395643.2 ENST00000395645.3 |
SLC5A10
|
solute carrier family 5 (sodium/sugar cotransporter), member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.0 | 0.3 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:2000690 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 1.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.2 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0070996 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |