Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for NKX2-8
Z-value: 0.68
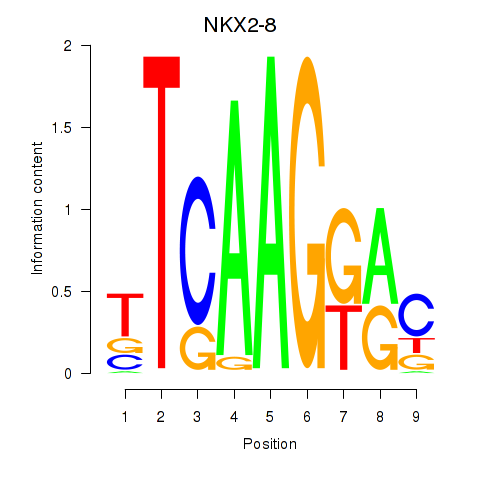
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.64 | 3.6e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_76251912 | 0.74 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_-_36779411 | 0.60 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr13_+_108921977 | 0.52 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_144679264 | 0.51 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr16_-_66952779 | 0.51 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_-_66952742 | 0.48 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr16_+_88872176 | 0.46 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr6_-_18249971 | 0.45 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr6_+_41010293 | 0.44 |
ENST00000373161.1
ENST00000373158.2 ENST00000470917.1 |
TSPO2
|
translocator protein 2 |
| chr1_+_228337553 | 0.40 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr6_+_34433844 | 0.38 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr3_-_187455680 | 0.33 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr4_-_164253738 | 0.33 |
ENST00000509586.1
ENST00000504391.1 ENST00000512819.1 |
NPY1R
|
neuropeptide Y receptor Y1 |
| chr4_+_141264597 | 0.33 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr2_-_55277692 | 0.33 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr7_-_99716914 | 0.32 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_143772060 | 0.32 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr6_+_43968306 | 0.32 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr6_+_125524785 | 0.31 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr17_+_42015654 | 0.31 |
ENST00000565120.1
|
RP11-527L4.2
|
Uncharacterized protein |
| chr7_-_156803329 | 0.30 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr18_+_13382553 | 0.30 |
ENST00000586222.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr22_-_21213676 | 0.30 |
ENST00000449120.1
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha |
| chr16_-_3350614 | 0.29 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr6_-_132272504 | 0.29 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr4_-_819901 | 0.29 |
ENST00000304062.6
|
CPLX1
|
complexin 1 |
| chr1_+_154401791 | 0.28 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr10_+_115674530 | 0.28 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr5_+_92919043 | 0.28 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr15_+_40453204 | 0.27 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr5_+_35852797 | 0.27 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr12_-_102591604 | 0.27 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chrX_-_64196351 | 0.27 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr16_+_58426296 | 0.26 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr14_+_58765103 | 0.25 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_136288740 | 0.25 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr1_+_109289279 | 0.25 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr5_+_176692466 | 0.24 |
ENST00000508029.1
ENST00000503056.1 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr8_+_104310661 | 0.23 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr8_-_125551278 | 0.23 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr17_+_76210267 | 0.22 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr3_+_187930719 | 0.22 |
ENST00000312675.4
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_77348821 | 0.21 |
ENST00000528364.1
ENST00000532069.1 ENST00000525428.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr10_-_118032697 | 0.21 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr14_-_58894223 | 0.21 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_+_55464600 | 0.20 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr5_-_78808617 | 0.20 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr19_+_44037546 | 0.20 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr7_-_99277610 | 0.20 |
ENST00000343703.5
ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr1_+_120839412 | 0.20 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr12_-_371994 | 0.20 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr14_-_92413353 | 0.19 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chrM_+_8366 | 0.19 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr18_-_44497308 | 0.19 |
ENST00000585916.1
ENST00000324794.7 ENST00000545673.1 |
PIAS2
|
protein inhibitor of activated STAT, 2 |
| chr4_-_110723134 | 0.19 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr11_-_77348796 | 0.19 |
ENST00000263309.3
ENST00000525064.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr2_-_55277654 | 0.19 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr16_+_28875268 | 0.18 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr9_+_21409146 | 0.18 |
ENST00000380205.1
|
IFNA8
|
interferon, alpha 8 |
| chr14_-_58894332 | 0.18 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr12_+_51442101 | 0.18 |
ENST00000550929.1
ENST00000262055.4 ENST00000550442.1 ENST00000549340.1 ENST00000548209.1 ENST00000548251.1 ENST00000550814.1 ENST00000547660.1 ENST00000380123.2 ENST00000548401.1 ENST00000418425.2 ENST00000547008.1 ENST00000552739.1 |
LETMD1
|
LETM1 domain containing 1 |
| chr14_-_58893876 | 0.17 |
ENST00000555097.1
ENST00000555404.1 |
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr7_-_5998714 | 0.17 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chrX_-_64196376 | 0.17 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr10_-_101673782 | 0.16 |
ENST00000422692.1
|
DNMBP
|
dynamin binding protein |
| chr7_-_86974767 | 0.16 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chr19_-_3600549 | 0.15 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr15_-_56757329 | 0.15 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr3_-_122134882 | 0.15 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr12_+_66218212 | 0.15 |
ENST00000393578.3
ENST00000425208.2 ENST00000536545.1 ENST00000354636.3 |
HMGA2
|
high mobility group AT-hook 2 |
| chr17_+_34901353 | 0.15 |
ENST00000593016.1
|
GGNBP2
|
gametogenetin binding protein 2 |
| chr14_+_37667230 | 0.15 |
ENST00000556451.1
ENST00000556753.1 ENST00000396294.2 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr5_+_34929677 | 0.15 |
ENST00000342382.4
ENST00000382021.2 ENST00000303525.7 |
DNAJC21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr16_+_226658 | 0.15 |
ENST00000320868.5
ENST00000397797.1 |
HBA1
|
hemoglobin, alpha 1 |
| chr20_-_17539456 | 0.14 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr16_+_89696692 | 0.14 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chrX_-_64196307 | 0.14 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr13_+_88325498 | 0.14 |
ENST00000400028.3
|
SLITRK5
|
SLIT and NTRK-like family, member 5 |
| chr17_-_46178527 | 0.13 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr16_+_28875126 | 0.13 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr22_+_31518938 | 0.13 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_42922173 | 0.13 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr11_-_10828892 | 0.13 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr2_-_55277436 | 0.12 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr14_+_21569245 | 0.12 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr19_+_2249308 | 0.12 |
ENST00000592877.1
ENST00000221496.4 |
AMH
|
anti-Mullerian hormone |
| chr5_-_176056974 | 0.12 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr13_-_46425865 | 0.12 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr2_-_3504587 | 0.12 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr6_+_84222220 | 0.11 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr3_-_8811288 | 0.11 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr17_-_33390667 | 0.11 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_+_149737202 | 0.11 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr4_-_70080449 | 0.11 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr14_-_24732368 | 0.11 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr1_+_173793777 | 0.11 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr7_+_112063192 | 0.11 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr16_+_67034456 | 0.10 |
ENST00000540579.1
|
CES4A
|
carboxylesterase 4A |
| chr10_-_118032979 | 0.10 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr12_+_49658855 | 0.10 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr12_+_16109519 | 0.10 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chrX_+_118533269 | 0.10 |
ENST00000336249.7
|
SLC25A43
|
solute carrier family 25, member 43 |
| chr10_-_14596140 | 0.10 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr3_-_114343039 | 0.10 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_110371720 | 0.10 |
ENST00000356688.4
|
SEPT10
|
septin 10 |
| chr5_-_95158644 | 0.10 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chrX_+_123094672 | 0.09 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr12_+_66218903 | 0.09 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chrX_-_72095622 | 0.09 |
ENST00000290273.5
|
DMRTC1
|
DMRT-like family C1 |
| chr19_-_17622269 | 0.09 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr2_-_152118352 | 0.09 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr5_+_80529104 | 0.09 |
ENST00000254035.4
ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr15_-_77363441 | 0.08 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr6_+_158957431 | 0.08 |
ENST00000367090.3
|
TMEM181
|
transmembrane protein 181 |
| chr4_-_170947565 | 0.08 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr14_-_24732403 | 0.08 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr13_+_46039037 | 0.08 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr16_+_29674540 | 0.08 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr7_+_75028199 | 0.08 |
ENST00000437796.1
|
TRIM73
|
tripartite motif containing 73 |
| chr2_+_11752379 | 0.08 |
ENST00000396123.1
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_-_58613323 | 0.08 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr14_+_76618242 | 0.08 |
ENST00000557542.1
ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L
|
G patch domain containing 2-like |
| chrX_+_72064690 | 0.07 |
ENST00000438696.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr6_+_84222194 | 0.07 |
ENST00000536636.1
|
PRSS35
|
protease, serine, 35 |
| chr12_+_121837905 | 0.07 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chr22_-_32341336 | 0.07 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr1_+_179050512 | 0.07 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr4_-_119274121 | 0.06 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr12_-_10320190 | 0.06 |
ENST00000543993.1
ENST00000339968.6 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr15_-_77363375 | 0.06 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr3_+_239652 | 0.06 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr17_-_46703826 | 0.06 |
ENST00000550387.1
ENST00000311177.5 |
HOXB9
|
homeobox B9 |
| chr8_-_141810634 | 0.06 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr6_-_32731299 | 0.06 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr11_+_59705928 | 0.06 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr3_-_87039662 | 0.06 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr1_-_220101944 | 0.05 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr3_-_53290016 | 0.05 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr17_+_73642486 | 0.05 |
ENST00000579469.1
|
SMIM6
|
small integral membrane protein 6 |
| chr7_+_73507409 | 0.05 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1 |
| chr11_+_64808675 | 0.05 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chrX_-_48858667 | 0.05 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr21_-_35899113 | 0.05 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr19_-_44259053 | 0.05 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr19_+_32836499 | 0.05 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chr8_-_145018905 | 0.05 |
ENST00000398774.2
|
PLEC
|
plectin |
| chr8_+_107593198 | 0.05 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr2_-_178129853 | 0.05 |
ENST00000397062.3
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr22_+_29834572 | 0.05 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr1_+_43291220 | 0.05 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr14_+_58765305 | 0.05 |
ENST00000445108.1
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr17_+_62503147 | 0.05 |
ENST00000553412.1
|
CEP95
|
centrosomal protein 95kDa |
| chrX_-_48776292 | 0.05 |
ENST00000376509.4
|
PIM2
|
pim-2 oncogene |
| chr7_+_135242652 | 0.05 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr5_-_179107975 | 0.04 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr18_+_44497455 | 0.04 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr11_+_35211429 | 0.04 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr6_-_137113604 | 0.04 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr4_-_100485143 | 0.04 |
ENST00000394877.3
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr9_-_99637820 | 0.04 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr7_+_30811004 | 0.04 |
ENST00000265299.6
|
FAM188B
|
family with sequence similarity 188, member B |
| chr2_-_55277512 | 0.04 |
ENST00000402434.2
|
RTN4
|
reticulon 4 |
| chr22_+_20877924 | 0.04 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr3_-_116163830 | 0.04 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chr17_+_73642315 | 0.04 |
ENST00000556126.2
|
SMIM6
|
small integral membrane protein 6 |
| chr7_-_95025661 | 0.04 |
ENST00000542556.1
ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1
PON3
|
paraoxonase 1 paraoxonase 3 |
| chr3_+_10183447 | 0.04 |
ENST00000345392.2
|
VHL
|
von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase |
| chr15_-_77363513 | 0.04 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr6_+_100054606 | 0.04 |
ENST00000369215.4
|
PRDM13
|
PR domain containing 13 |
| chr11_+_18720316 | 0.03 |
ENST00000280734.2
|
TMEM86A
|
transmembrane protein 86A |
| chrX_+_77154935 | 0.03 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr9_-_128412696 | 0.03 |
ENST00000420643.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr19_-_51893782 | 0.03 |
ENST00000570516.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr19_-_51893827 | 0.03 |
ENST00000574814.1
|
CTD-2616J11.4
|
chromosome 19 open reading frame 84 |
| chr10_+_123951957 | 0.03 |
ENST00000514539.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr8_-_29120604 | 0.03 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr15_+_45422178 | 0.03 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr22_+_32754139 | 0.03 |
ENST00000382088.3
|
RFPL3
|
ret finger protein-like 3 |
| chr12_-_75603482 | 0.03 |
ENST00000341669.3
ENST00000298972.1 ENST00000350228.2 |
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_-_133429091 | 0.03 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr10_+_24755416 | 0.03 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr22_+_32753854 | 0.03 |
ENST00000249007.4
|
RFPL3
|
ret finger protein-like 3 |
| chrX_+_115567767 | 0.03 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr4_-_100485183 | 0.03 |
ENST00000394876.2
|
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr12_-_49504449 | 0.03 |
ENST00000547675.1
|
LMBR1L
|
limb development membrane protein 1-like |
| chr5_+_43602750 | 0.03 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr3_-_196242233 | 0.03 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr17_+_39994032 | 0.03 |
ENST00000293303.4
ENST00000438813.1 |
KLHL10
|
kelch-like family member 10 |
| chr14_+_37667118 | 0.03 |
ENST00000556615.1
ENST00000327441.7 ENST00000536774.1 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr4_-_153601136 | 0.03 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr17_-_28661065 | 0.02 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr20_-_43936937 | 0.02 |
ENST00000342716.4
ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4
|
matrilin 4 |
| chr13_+_108922228 | 0.02 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr4_+_76481258 | 0.02 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr17_-_62503015 | 0.02 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr7_-_137686791 | 0.02 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr9_+_109685630 | 0.02 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr11_+_71791849 | 0.02 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr12_+_66218598 | 0.02 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr3_-_178103144 | 0.02 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.4 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.3 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0043605 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0002884 | negative regulation of type IV hypersensitivity(GO:0001808) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:2001256 | chemical homeostasis within a tissue(GO:0048875) regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.0 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.3 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.3 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.3 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.4 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |