Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
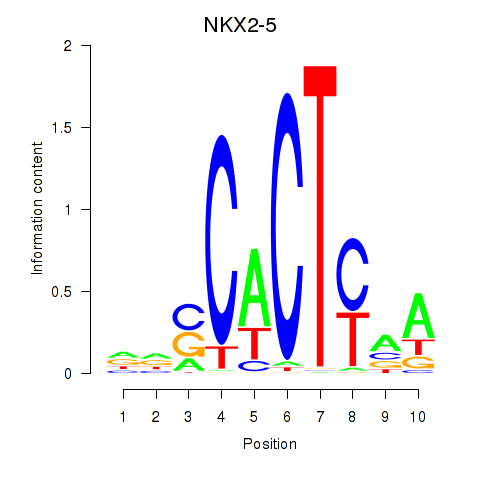
Results for NKX2-5
Z-value: 0.80
Transcription factors associated with NKX2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-5
|
ENSG00000183072.9 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-5 | hg19_v2_chr5_-_172662197_172662216 | 0.52 | 4.8e-01 | Click! |
Activity profile of NKX2-5 motif
Sorted Z-values of NKX2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_115567767 | 0.40 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr12_-_123717711 | 0.35 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr1_+_150254936 | 0.33 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr8_-_73793975 | 0.32 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr1_-_11115877 | 0.31 |
ENST00000490101.1
|
SRM
|
spermidine synthase |
| chr17_+_74261413 | 0.30 |
ENST00000587913.1
|
UBALD2
|
UBA-like domain containing 2 |
| chr20_-_56285595 | 0.30 |
ENST00000395816.3
ENST00000347215.4 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr4_-_168155730 | 0.29 |
ENST00000502330.1
ENST00000357154.3 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_113354341 | 0.28 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr16_+_77233294 | 0.28 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr1_+_26606608 | 0.27 |
ENST00000319041.6
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr7_-_27224795 | 0.27 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr7_-_99774945 | 0.26 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr17_-_29641104 | 0.26 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr17_-_55911970 | 0.26 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr7_-_94953878 | 0.25 |
ENST00000222381.3
|
PON1
|
paraoxonase 1 |
| chr4_+_81118647 | 0.23 |
ENST00000415738.2
|
PRDM8
|
PR domain containing 8 |
| chr19_+_39926791 | 0.23 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr6_+_26273144 | 0.22 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr13_-_41240717 | 0.22 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr9_-_138393580 | 0.20 |
ENST00000371791.1
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr6_-_41715128 | 0.20 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr5_-_138730817 | 0.20 |
ENST00000434752.2
|
PROB1
|
proline-rich basic protein 1 |
| chr9_-_25678856 | 0.20 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr7_-_107883678 | 0.20 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr18_+_42260059 | 0.20 |
ENST00000426838.4
|
SETBP1
|
SET binding protein 1 |
| chr10_-_49813090 | 0.19 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr19_-_5680891 | 0.19 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr3_-_194072019 | 0.19 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr7_+_129007964 | 0.18 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr8_+_9009296 | 0.18 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chrX_-_14047996 | 0.18 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr1_-_202858227 | 0.18 |
ENST00000367262.3
|
RABIF
|
RAB interacting factor |
| chr15_+_42696954 | 0.18 |
ENST00000337571.4
ENST00000569136.1 |
CAPN3
|
calpain 3, (p94) |
| chr14_+_78227105 | 0.17 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chrX_-_55057403 | 0.17 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr21_+_44313375 | 0.17 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr9_+_35906176 | 0.17 |
ENST00000354323.2
|
HRCT1
|
histidine rich carboxyl terminus 1 |
| chr18_+_42277095 | 0.16 |
ENST00000591940.1
|
SETBP1
|
SET binding protein 1 |
| chr16_-_30134524 | 0.16 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr7_-_27224842 | 0.16 |
ENST00000517402.1
|
HOXA11
|
homeobox A11 |
| chr17_+_74261277 | 0.16 |
ENST00000327490.6
|
UBALD2
|
UBA-like domain containing 2 |
| chr4_-_168155577 | 0.16 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr14_+_105927191 | 0.15 |
ENST00000550551.1
|
MTA1
|
metastasis associated 1 |
| chr17_-_7493390 | 0.15 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr18_+_61575200 | 0.15 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_+_108746282 | 0.14 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr17_-_29641084 | 0.14 |
ENST00000544462.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr3_-_11685345 | 0.14 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr7_-_122339162 | 0.14 |
ENST00000340112.2
|
RNF133
|
ring finger protein 133 |
| chr19_+_14672755 | 0.14 |
ENST00000594545.1
|
TECR
|
trans-2,3-enoyl-CoA reductase |
| chr15_+_42697018 | 0.13 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr16_-_30134266 | 0.13 |
ENST00000484663.1
ENST00000478356.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr2_+_219646462 | 0.13 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr7_-_128415844 | 0.13 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr4_-_2264015 | 0.13 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chrX_-_49041242 | 0.13 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr5_+_75699040 | 0.13 |
ENST00000274364.6
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr20_+_55841819 | 0.12 |
ENST00000412321.1
ENST00000426580.1 |
RP4-813D12.3
|
RP4-813D12.3 |
| chr3_-_123710893 | 0.12 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr4_+_110769258 | 0.12 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr2_-_27545431 | 0.12 |
ENST00000233545.2
|
MPV17
|
MpV17 mitochondrial inner membrane protein |
| chr1_+_76540386 | 0.12 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr22_-_38577731 | 0.12 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr10_+_86184676 | 0.12 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
| chr10_-_27529486 | 0.11 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr7_-_92146729 | 0.11 |
ENST00000541751.1
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr10_-_128359074 | 0.11 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr6_-_134373732 | 0.11 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr22_-_38577782 | 0.11 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr9_+_17579084 | 0.11 |
ENST00000380607.4
|
SH3GL2
|
SH3-domain GRB2-like 2 |
| chr3_+_23847394 | 0.11 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr5_+_74062806 | 0.10 |
ENST00000296802.5
|
NSA2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr1_-_149982624 | 0.10 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr2_+_201997492 | 0.10 |
ENST00000494258.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr12_-_123717643 | 0.10 |
ENST00000541437.1
ENST00000606320.1 |
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr12_+_93772402 | 0.10 |
ENST00000546925.1
|
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr8_-_49833978 | 0.10 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr11_-_118436606 | 0.10 |
ENST00000530872.1
|
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr5_+_122110691 | 0.10 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr1_+_161676739 | 0.09 |
ENST00000236938.6
ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA
|
Fc receptor-like A |
| chr17_-_56621665 | 0.09 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr6_+_31916733 | 0.09 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr8_+_132916318 | 0.09 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr6_-_94129244 | 0.09 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr12_-_50222187 | 0.09 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr8_-_10588010 | 0.09 |
ENST00000304501.1
|
SOX7
|
SRY (sex determining region Y)-box 7 |
| chr17_-_3500392 | 0.08 |
ENST00000571088.1
ENST00000174621.6 |
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr12_+_14518598 | 0.08 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr2_-_201936302 | 0.08 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr7_-_38969150 | 0.08 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr6_+_33378738 | 0.08 |
ENST00000374512.3
ENST00000374516.3 |
PHF1
|
PHD finger protein 1 |
| chr8_-_49834299 | 0.08 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_-_371994 | 0.07 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr1_+_207494853 | 0.07 |
ENST00000367064.3
ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55
|
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr17_-_46667594 | 0.07 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr7_-_81399355 | 0.07 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr11_+_64879317 | 0.07 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr2_-_85895295 | 0.07 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chr7_-_81399329 | 0.07 |
ENST00000453411.1
ENST00000444829.2 |
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_-_113160826 | 0.07 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr8_+_145729465 | 0.07 |
ENST00000394955.2
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase) |
| chr3_+_125687987 | 0.06 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chrX_+_86772787 | 0.06 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chrX_+_40944871 | 0.06 |
ENST00000378308.2
ENST00000324545.8 |
USP9X
|
ubiquitin specific peptidase 9, X-linked |
| chr8_-_72459885 | 0.06 |
ENST00000523987.1
|
RP11-1102P16.1
|
Uncharacterized protein |
| chr14_+_64565442 | 0.06 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr5_+_134181625 | 0.06 |
ENST00000394976.3
|
C5orf24
|
chromosome 5 open reading frame 24 |
| chr5_-_74062867 | 0.06 |
ENST00000509097.1
|
GFM2
|
G elongation factor, mitochondrial 2 |
| chr11_+_94277017 | 0.06 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr2_+_136289030 | 0.06 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr7_-_81399287 | 0.06 |
ENST00000354224.6
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_+_81589254 | 0.06 |
ENST00000394652.2
|
IL16
|
interleukin 16 |
| chr16_+_69458537 | 0.06 |
ENST00000515314.1
ENST00000561792.1 ENST00000568237.1 |
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane) |
| chr10_-_99531709 | 0.06 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr22_-_29137771 | 0.06 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr4_+_156680518 | 0.06 |
ENST00000513437.1
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr1_+_161676983 | 0.05 |
ENST00000367957.2
|
FCRLA
|
Fc receptor-like A |
| chr1_+_100817262 | 0.05 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr6_-_112115074 | 0.05 |
ENST00000368667.2
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr15_-_42840961 | 0.05 |
ENST00000563454.1
ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57
|
leucine rich repeat containing 57 |
| chr4_-_168155700 | 0.05 |
ENST00000357545.4
ENST00000512648.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr7_-_81399411 | 0.05 |
ENST00000423064.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr22_-_50524298 | 0.05 |
ENST00000311597.5
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr16_+_8814563 | 0.05 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr14_-_75179774 | 0.05 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr16_-_70729496 | 0.05 |
ENST00000567648.1
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr1_-_23520755 | 0.05 |
ENST00000314113.3
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr20_+_57226284 | 0.05 |
ENST00000458280.1
ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16
|
syntaxin 16 |
| chr14_+_81421861 | 0.05 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr10_+_111967345 | 0.05 |
ENST00000332674.5
ENST00000453116.1 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr2_-_180610767 | 0.05 |
ENST00000409343.1
|
ZNF385B
|
zinc finger protein 385B |
| chr15_-_60683326 | 0.05 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr17_+_29421987 | 0.05 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr6_+_30297306 | 0.05 |
ENST00000420746.1
ENST00000513556.1 |
TRIM39
TRIM39-RPP21
|
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr1_-_36947120 | 0.05 |
ENST00000361632.4
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr22_-_50523760 | 0.05 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr16_+_765092 | 0.05 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr15_+_74421961 | 0.05 |
ENST00000565159.1
ENST00000567206.1 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chrX_+_14547632 | 0.05 |
ENST00000218075.4
|
GLRA2
|
glycine receptor, alpha 2 |
| chr16_-_30134441 | 0.05 |
ENST00000395200.1
|
MAPK3
|
mitogen-activated protein kinase 3 |
| chr2_+_201936458 | 0.05 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr13_+_109248500 | 0.04 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr5_+_34685805 | 0.04 |
ENST00000508315.1
|
RAI14
|
retinoic acid induced 14 |
| chr10_+_133753533 | 0.04 |
ENST00000422256.2
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr1_+_202431859 | 0.04 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_+_57769635 | 0.04 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr1_-_115259337 | 0.04 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr16_-_3149278 | 0.04 |
ENST00000575108.1
ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr17_-_40169429 | 0.04 |
ENST00000316603.7
ENST00000588641.1 |
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr17_-_8279977 | 0.04 |
ENST00000396267.1
|
KRBA2
|
KRAB-A domain containing 2 |
| chr11_+_18417813 | 0.04 |
ENST00000540430.1
ENST00000379412.5 |
LDHA
|
lactate dehydrogenase A |
| chr6_+_109169591 | 0.04 |
ENST00000368972.3
ENST00000392644.4 |
ARMC2
|
armadillo repeat containing 2 |
| chr8_+_133879193 | 0.04 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr10_+_79626633 | 0.04 |
ENST00000372387.1
|
AL391421.1
|
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
| chr14_+_81421355 | 0.04 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr20_-_271009 | 0.04 |
ENST00000382369.5
|
C20orf96
|
chromosome 20 open reading frame 96 |
| chr6_-_112115103 | 0.04 |
ENST00000462598.3
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr11_-_118436707 | 0.03 |
ENST00000264020.2
ENST00000264021.3 |
IFT46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr4_-_168155417 | 0.03 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chrX_-_13835147 | 0.03 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr11_-_108464465 | 0.03 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr15_+_78799948 | 0.03 |
ENST00000566332.1
|
HYKK
|
hydroxylysine kinase |
| chrX_-_154299501 | 0.03 |
ENST00000369476.3
ENST00000369484.3 |
MTCP1
CMC4
|
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chrX_-_49965663 | 0.03 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chr17_+_40811283 | 0.03 |
ENST00000251412.7
|
TUBG2
|
tubulin, gamma 2 |
| chr19_+_5681153 | 0.03 |
ENST00000579559.1
ENST00000577222.1 |
HSD11B1L
RPL36
|
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr19_-_5680499 | 0.03 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr4_-_74853897 | 0.03 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr20_+_57226841 | 0.03 |
ENST00000358029.4
ENST00000361830.3 |
STX16
|
syntaxin 16 |
| chr17_+_58755184 | 0.02 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr1_+_170115142 | 0.02 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr11_+_17756279 | 0.02 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr17_-_13505219 | 0.02 |
ENST00000284110.1
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
| chr7_-_81399438 | 0.02 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr1_-_160040038 | 0.02 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr18_-_13915530 | 0.02 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr17_-_40169659 | 0.02 |
ENST00000457167.4
|
DNAJC7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr8_-_120651020 | 0.02 |
ENST00000522826.1
ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr1_-_55352834 | 0.02 |
ENST00000371269.3
|
DHCR24
|
24-dehydrocholesterol reductase |
| chr1_+_159796534 | 0.02 |
ENST00000289707.5
|
SLAMF8
|
SLAM family member 8 |
| chr12_-_53097247 | 0.02 |
ENST00000341809.3
ENST00000537195.1 |
KRT77
|
keratin 77 |
| chr3_+_45984947 | 0.02 |
ENST00000304552.4
|
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr1_+_56880606 | 0.02 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr17_+_45973516 | 0.02 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr15_-_74726283 | 0.02 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr11_-_18813110 | 0.02 |
ENST00000396168.1
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chr1_+_154301264 | 0.01 |
ENST00000341822.2
|
ATP8B2
|
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr11_+_120973375 | 0.01 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr12_+_111051832 | 0.01 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr6_-_27279949 | 0.01 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr1_+_203765437 | 0.01 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr19_-_45873642 | 0.01 |
ENST00000485403.2
ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr3_-_187388173 | 0.01 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr1_-_207143802 | 0.01 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr17_+_66521936 | 0.01 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_+_144354644 | 0.01 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr11_+_18417948 | 0.01 |
ENST00000542179.1
|
LDHA
|
lactate dehydrogenase A |
| chrX_+_71353499 | 0.01 |
ENST00000373677.1
|
NHSL2
|
NHS-like 2 |
| chr9_+_118950325 | 0.01 |
ENST00000534838.1
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.1 | 0.2 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 0.2 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.4 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:1902617 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.0 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.4 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0044035 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |