Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for NKX2-4
Z-value: 1.52
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.4 | NK2 homeobox 4 |
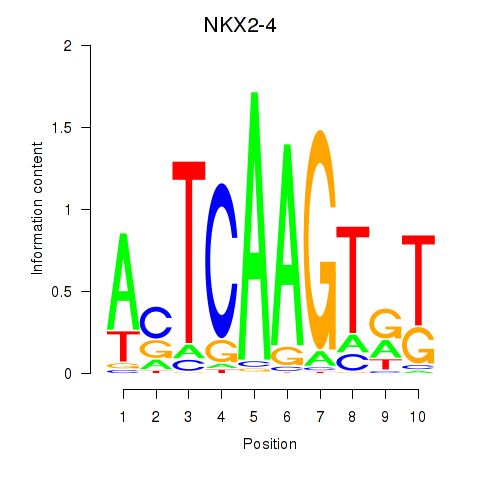
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_38177575 | 1.41 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr14_+_32414059 | 1.30 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr4_-_155533787 | 1.00 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr11_-_104035088 | 0.98 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr16_+_12059091 | 0.98 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr3_+_118905564 | 0.78 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr4_+_72204755 | 0.75 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr19_-_40440533 | 0.75 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr4_+_155484155 | 0.75 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_90033968 | 0.74 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr12_-_71533055 | 0.73 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr2_+_109065634 | 0.73 |
ENST00000409821.1
|
GCC2
|
GRIP and coiled-coil domain containing 2 |
| chr6_-_73935163 | 0.72 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr1_+_95583479 | 0.70 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr3_-_49066811 | 0.70 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr3_-_112329110 | 0.67 |
ENST00000479368.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr1_-_197036364 | 0.65 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_-_36779411 | 0.62 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr17_-_38574169 | 0.62 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr10_-_18944123 | 0.61 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr17_-_19062424 | 0.60 |
ENST00000399083.1
|
AC007952.6
|
Uncharacterized protein |
| chr16_+_3062457 | 0.59 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr18_+_9136758 | 0.58 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr22_+_25465786 | 0.54 |
ENST00000401395.1
|
KIAA1671
|
KIAA1671 |
| chr2_+_120687335 | 0.54 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr2_-_85828867 | 0.53 |
ENST00000425160.1
|
TMEM150A
|
transmembrane protein 150A |
| chr10_+_99332529 | 0.52 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr12_+_123237321 | 0.51 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr15_+_75315896 | 0.50 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr1_+_61869748 | 0.49 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr13_-_43566301 | 0.49 |
ENST00000398762.3
ENST00000313640.7 ENST00000313624.7 |
EPSTI1
|
epithelial stromal interaction 1 (breast) |
| chrX_-_84363974 | 0.48 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr15_+_34394257 | 0.47 |
ENST00000397766.2
|
PGBD4
|
piggyBac transposable element derived 4 |
| chr9_-_3469181 | 0.47 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr11_+_107992518 | 0.47 |
ENST00000527942.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr2_+_161993465 | 0.47 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr6_+_27925019 | 0.46 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr7_+_134551583 | 0.46 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr2_-_220025263 | 0.45 |
ENST00000457600.1
|
NHEJ1
|
nonhomologous end-joining factor 1 |
| chr21_+_40177143 | 0.45 |
ENST00000360214.3
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr11_-_506316 | 0.45 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr14_+_64680854 | 0.45 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_-_12503156 | 0.45 |
ENST00000543314.1
ENST00000396349.3 |
MANSC1
|
MANSC domain containing 1 |
| chr7_+_65552756 | 0.44 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr11_-_105892937 | 0.44 |
ENST00000301919.4
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr7_-_17980091 | 0.44 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr15_-_31523036 | 0.43 |
ENST00000559094.1
ENST00000558388.2 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr2_+_53994927 | 0.43 |
ENST00000295304.4
|
CHAC2
|
ChaC, cation transport regulator homolog 2 (E. coli) |
| chrX_+_70443050 | 0.43 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr11_+_33037652 | 0.42 |
ENST00000311388.3
|
DEPDC7
|
DEP domain containing 7 |
| chr14_-_23451845 | 0.41 |
ENST00000262713.2
|
AJUBA
|
ajuba LIM protein |
| chr3_+_30647994 | 0.41 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr5_+_175298573 | 0.40 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr11_+_61976137 | 0.40 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr17_+_4634705 | 0.40 |
ENST00000575284.1
ENST00000573708.1 ENST00000293777.5 |
MED11
|
mediator complex subunit 11 |
| chr9_-_86432547 | 0.39 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr12_-_58329819 | 0.39 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr6_+_42018251 | 0.39 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr18_-_28622699 | 0.38 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr19_-_42758040 | 0.38 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr15_+_36887069 | 0.38 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr4_-_100242549 | 0.37 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr1_-_89458287 | 0.37 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr19_-_12444491 | 0.37 |
ENST00000293725.5
|
ZNF563
|
zinc finger protein 563 |
| chr18_-_28622774 | 0.37 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr9_+_140119618 | 0.37 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr1_+_161136180 | 0.36 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr15_-_63448973 | 0.36 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr17_-_5138099 | 0.35 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr4_+_3388057 | 0.35 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr14_-_81408093 | 0.35 |
ENST00000555265.1
|
CEP128
|
centrosomal protein 128kDa |
| chr4_-_682769 | 0.34 |
ENST00000507165.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr6_-_149806105 | 0.34 |
ENST00000389942.5
ENST00000416573.2 ENST00000542614.1 ENST00000409806.3 |
ZC3H12D
|
zinc finger CCCH-type containing 12D |
| chr14_-_23451467 | 0.34 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr1_+_64239657 | 0.34 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr8_+_97597148 | 0.34 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr7_-_86848933 | 0.34 |
ENST00000423734.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr7_-_55606346 | 0.33 |
ENST00000545390.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr2_+_205410723 | 0.33 |
ENST00000358768.2
ENST00000351153.1 ENST00000349953.3 |
PARD3B
|
par-3 family cell polarity regulator beta |
| chr6_-_28303901 | 0.33 |
ENST00000439158.1
ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr9_-_36258326 | 0.33 |
ENST00000447283.2
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr12_-_102591604 | 0.33 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr5_-_78808617 | 0.32 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_96012614 | 0.32 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr1_+_196788887 | 0.32 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr3_+_186158169 | 0.31 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chrX_-_154563889 | 0.31 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr1_-_70671216 | 0.31 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr14_-_96830207 | 0.31 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr12_+_27863706 | 0.31 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr12_+_50451331 | 0.31 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr17_+_19122674 | 0.31 |
ENST00000436914.1
|
AC106017.1
|
Uncharacterized protein |
| chr2_-_3521518 | 0.31 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr4_-_68411275 | 0.31 |
ENST00000273853.6
|
CENPC
|
centromere protein C |
| chr12_+_50451462 | 0.31 |
ENST00000447966.2
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr8_-_65730127 | 0.31 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr7_-_102252589 | 0.30 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr19_+_41856816 | 0.30 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr22_-_18923655 | 0.30 |
ENST00000438924.1
ENST00000457083.1 ENST00000420436.1 ENST00000334029.2 ENST00000357068.6 |
PRODH
|
proline dehydrogenase (oxidase) 1 |
| chr6_+_37225540 | 0.30 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr4_+_169418195 | 0.29 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_18996287 | 0.29 |
ENST00000399091.1
ENST00000443876.1 ENST00000428928.1 |
AC007952.5
|
Uncharacterized protein ENSP00000382042 |
| chr3_+_189349162 | 0.29 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr8_+_104310661 | 0.29 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr15_+_42694573 | 0.29 |
ENST00000397200.4
ENST00000569827.1 |
CAPN3
|
calpain 3, (p94) |
| chr14_+_96829886 | 0.29 |
ENST00000556095.1
|
GSKIP
|
GSK3B interacting protein |
| chr11_-_44972418 | 0.29 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr6_+_26158343 | 0.29 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chr11_+_35965531 | 0.28 |
ENST00000528989.1
ENST00000524419.1 ENST00000315571.5 |
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3 |
| chr12_+_133757995 | 0.28 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr11_+_34642656 | 0.28 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr11_-_44972476 | 0.28 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr3_+_148847371 | 0.27 |
ENST00000296051.2
ENST00000460120.1 |
HPS3
|
Hermansky-Pudlak syndrome 3 |
| chr5_-_42811986 | 0.27 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_-_86849025 | 0.27 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr17_+_46918925 | 0.27 |
ENST00000502761.1
|
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr8_-_141774467 | 0.27 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr16_-_31161380 | 0.27 |
ENST00000569305.1
ENST00000418068.2 ENST00000268281.4 |
PRSS36
|
protease, serine, 36 |
| chr17_+_66244071 | 0.27 |
ENST00000580548.1
ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr17_-_63822563 | 0.27 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr7_-_54826869 | 0.26 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr16_-_12062333 | 0.26 |
ENST00000597717.1
|
AC007216.2
|
Uncharacterized protein |
| chr14_-_105708942 | 0.26 |
ENST00000549655.1
|
BRF1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr9_-_99616642 | 0.26 |
ENST00000478850.1
ENST00000481138.1 |
ZNF782
|
zinc finger protein 782 |
| chr3_-_88108192 | 0.26 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_-_85581623 | 0.26 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr3_+_159706537 | 0.26 |
ENST00000305579.2
ENST00000480787.1 ENST00000466512.1 |
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr15_-_83474806 | 0.26 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr2_-_225434538 | 0.26 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr8_-_74206673 | 0.26 |
ENST00000396465.1
|
RPL7
|
ribosomal protein L7 |
| chr10_-_115423792 | 0.26 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr1_-_12908578 | 0.26 |
ENST00000317869.6
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1 |
| chr12_-_39837192 | 0.26 |
ENST00000361961.3
ENST00000395670.3 |
KIF21A
|
kinesin family member 21A |
| chr19_+_46531127 | 0.25 |
ENST00000601033.1
|
CTC-344H19.4
|
CTC-344H19.4 |
| chr4_+_86749045 | 0.25 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_60094735 | 0.25 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr16_-_31100284 | 0.25 |
ENST00000280606.6
|
PRSS53
|
protease, serine, 53 |
| chr14_+_56078695 | 0.25 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr8_+_22429205 | 0.25 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr2_+_145425534 | 0.25 |
ENST00000432608.1
ENST00000597655.1 ENST00000598659.1 ENST00000600679.1 ENST00000601277.1 ENST00000451027.1 ENST00000445791.1 ENST00000596540.1 ENST00000596230.1 ENST00000594471.1 ENST00000598248.1 ENST00000597469.1 ENST00000431734.1 ENST00000595686.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr17_-_15168624 | 0.25 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr2_+_201677390 | 0.25 |
ENST00000447069.1
|
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr17_+_57297807 | 0.25 |
ENST00000284116.4
ENST00000581140.1 ENST00000581276.1 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr12_+_124069070 | 0.25 |
ENST00000262225.3
ENST00000438031.2 |
TMED2
|
transmembrane emp24 domain trafficking protein 2 |
| chr6_+_35265586 | 0.24 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr13_+_53216565 | 0.24 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr17_+_75277492 | 0.24 |
ENST00000427177.1
ENST00000591198.1 |
SEPT9
|
septin 9 |
| chr5_+_154135453 | 0.24 |
ENST00000517616.1
ENST00000518892.1 |
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr11_+_34999328 | 0.24 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr11_-_14521349 | 0.24 |
ENST00000534234.1
|
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr2_+_36923830 | 0.24 |
ENST00000379242.3
ENST00000389975.3 |
VIT
|
vitrin |
| chr11_-_44972390 | 0.24 |
ENST00000395648.3
ENST00000531928.2 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr2_+_179316163 | 0.24 |
ENST00000409117.3
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr5_-_34919094 | 0.23 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr14_+_24702099 | 0.23 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr19_-_20748614 | 0.23 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr2_+_37571845 | 0.23 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr2_+_103378472 | 0.23 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr7_-_92463210 | 0.23 |
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6 |
| chr3_-_56950407 | 0.23 |
ENST00000496106.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr14_+_24702073 | 0.22 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr1_-_115301235 | 0.22 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr10_+_52094298 | 0.22 |
ENST00000595931.1
|
AC069547.1
|
HCG1745369; PRO3073; Uncharacterized protein |
| chr9_+_131445928 | 0.22 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr5_+_139505520 | 0.22 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr11_+_93479588 | 0.22 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr16_+_524850 | 0.22 |
ENST00000450428.1
ENST00000452814.1 |
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr11_-_4719072 | 0.22 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr1_-_160492994 | 0.21 |
ENST00000368055.1
ENST00000368057.3 ENST00000368059.3 |
SLAMF6
|
SLAM family member 6 |
| chr17_-_19648416 | 0.21 |
ENST00000426645.2
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr5_-_68339648 | 0.21 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr12_-_104532062 | 0.21 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr19_+_41620335 | 0.21 |
ENST00000331105.2
|
CYP2F1
|
cytochrome P450, family 2, subfamily F, polypeptide 1 |
| chr12_+_58176525 | 0.21 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr9_+_124048864 | 0.21 |
ENST00000545652.1
|
GSN
|
gelsolin |
| chr17_-_62493131 | 0.21 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr12_-_7261772 | 0.20 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr3_-_69249863 | 0.20 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr17_-_15587602 | 0.20 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr3_+_101504200 | 0.20 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr5_+_175298674 | 0.20 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr7_-_86849883 | 0.20 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr4_-_175443943 | 0.20 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr8_+_145159846 | 0.20 |
ENST00000532522.1
ENST00000527572.1 ENST00000527058.1 |
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chr14_+_24702127 | 0.20 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr6_-_136610911 | 0.20 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr19_+_59055412 | 0.19 |
ENST00000593582.1
|
TRIM28
|
tripartite motif containing 28 |
| chr16_-_3137080 | 0.19 |
ENST00000574387.1
ENST00000571404.1 |
RP11-473M20.9
|
RP11-473M20.9 |
| chr2_-_105953912 | 0.19 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chrX_+_49091920 | 0.19 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr17_-_34329084 | 0.19 |
ENST00000354059.4
ENST00000536149.1 |
CCL15
CCL14
|
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr17_-_37844267 | 0.19 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr3_+_156807663 | 0.19 |
ENST00000467995.1
ENST00000474477.1 ENST00000471719.1 |
LINC00881
|
long intergenic non-protein coding RNA 881 |
| chr2_+_161993412 | 0.19 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_104909435 | 0.19 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr14_+_24701819 | 0.19 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr16_+_57496299 | 0.18 |
ENST00000219252.5
|
POLR2C
|
polymerase (RNA) II (DNA directed) polypeptide C, 33kDa |
| chr2_-_103353277 | 0.18 |
ENST00000258436.5
|
MFSD9
|
major facilitator superfamily domain containing 9 |
| chr11_-_64856497 | 0.18 |
ENST00000524632.1
ENST00000530719.1 |
AP003068.6
|
transmembrane protein 262 |
| chr12_+_133758115 | 0.18 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.2 | 0.7 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 0.5 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.2 | 0.5 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.4 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.1 | GO:0044268 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.5 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.2 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.7 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.8 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.2 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.3 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0034255 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.3 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 1.1 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.3 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.7 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) tRNA transcription(GO:0009304) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.4 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.2 | GO:1904628 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) negative regulation of monocyte differentiation(GO:0045656) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.3 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.2 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 1.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.9 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.2 | 1.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.3 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.1 | 0.3 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.2 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |