Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
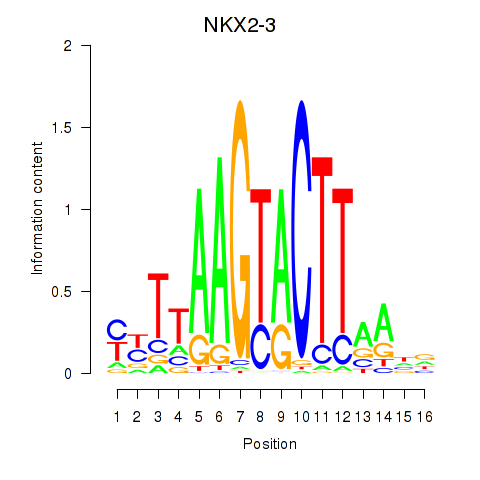
Results for NKX2-3
Z-value: 0.87
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-3 | hg19_v2_chr10_+_101292684_101292706 | -0.09 | 9.1e-01 | Click! |
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_65266507 | 1.01 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr3_+_149192475 | 0.84 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr16_-_66952779 | 0.60 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr3_+_149191723 | 0.59 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr15_-_56757329 | 0.55 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr16_-_66952742 | 0.53 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chrX_+_69509870 | 0.46 |
ENST00000374388.3
|
KIF4A
|
kinesin family member 4A |
| chr9_-_116840728 | 0.43 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr5_+_150404904 | 0.38 |
ENST00000521632.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr14_+_94547628 | 0.37 |
ENST00000555523.1
|
IFI27L1
|
interferon, alpha-inducible protein 27-like 1 |
| chr14_+_100531738 | 0.36 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr14_+_100531615 | 0.35 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr20_+_32782375 | 0.35 |
ENST00000568305.1
|
ASIP
|
agouti signaling protein |
| chr2_-_239198743 | 0.34 |
ENST00000440245.1
ENST00000431832.1 |
PER2
|
period circadian clock 2 |
| chr16_-_69166031 | 0.34 |
ENST00000398235.2
ENST00000520529.1 |
CHTF8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr18_-_43684230 | 0.34 |
ENST00000592989.1
ENST00000589869.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr19_-_17622269 | 0.33 |
ENST00000595116.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr18_-_25616519 | 0.33 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr18_-_53735601 | 0.32 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr4_-_100242549 | 0.32 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr16_+_53412368 | 0.32 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr7_-_29186008 | 0.31 |
ENST00000396276.3
ENST00000265394.5 |
CPVL
|
carboxypeptidase, vitellogenic-like |
| chr17_+_26662679 | 0.30 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr10_-_32345305 | 0.28 |
ENST00000302418.4
|
KIF5B
|
kinesin family member 5B |
| chr1_+_205682497 | 0.28 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr7_-_84569561 | 0.28 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr7_+_7196565 | 0.27 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr12_-_52604607 | 0.27 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr11_+_62379194 | 0.27 |
ENST00000525801.1
ENST00000534093.1 |
ROM1
|
retinal outer segment membrane protein 1 |
| chr6_+_31126291 | 0.27 |
ENST00000376257.3
ENST00000376255.4 |
TCF19
|
transcription factor 19 |
| chr3_+_30648066 | 0.26 |
ENST00000359013.4
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr8_+_41386761 | 0.26 |
ENST00000523277.2
|
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr1_-_197036364 | 0.25 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr19_-_39368887 | 0.25 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr10_+_99332198 | 0.24 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr3_+_57094469 | 0.24 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr3_+_30647994 | 0.24 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr16_+_32264040 | 0.24 |
ENST00000398664.3
|
TP53TG3D
|
TP53 target 3D |
| chr20_+_56964253 | 0.24 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr13_+_50570019 | 0.23 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr2_+_120687335 | 0.23 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr2_+_204103663 | 0.23 |
ENST00000356079.4
ENST00000429815.2 |
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr14_+_94547675 | 0.23 |
ENST00000393115.3
ENST00000554166.1 ENST00000556381.1 ENST00000553664.1 ENST00000555341.1 ENST00000557218.1 ENST00000554544.1 ENST00000557066.1 |
IFI27L1
|
interferon, alpha-inducible protein 27-like 1 |
| chr2_+_114163945 | 0.23 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr10_-_5227096 | 0.22 |
ENST00000488756.1
ENST00000334314.3 |
AKR1CL1
|
aldo-keto reductase family 1, member C-like 1 |
| chr19_+_14492217 | 0.22 |
ENST00000587606.1
ENST00000586517.1 ENST00000591080.1 |
CD97
|
CD97 molecule |
| chr15_+_40674920 | 0.22 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr10_+_103348031 | 0.22 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr1_+_43824669 | 0.22 |
ENST00000372462.1
|
CDC20
|
cell division cycle 20 |
| chr1_+_65613217 | 0.22 |
ENST00000545314.1
|
AK4
|
adenylate kinase 4 |
| chr13_-_46679144 | 0.22 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr10_+_1120312 | 0.22 |
ENST00000436154.1
|
WDR37
|
WD repeat domain 37 |
| chr22_+_44427230 | 0.21 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr13_-_46679185 | 0.21 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr20_-_1317555 | 0.21 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr11_+_64950801 | 0.21 |
ENST00000526468.1
|
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr16_+_68877496 | 0.21 |
ENST00000261778.1
|
TANGO6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr11_-_64856497 | 0.21 |
ENST00000524632.1
ENST00000530719.1 |
AP003068.6
|
transmembrane protein 262 |
| chr1_-_26633480 | 0.20 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chrX_+_153029633 | 0.20 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr18_-_43684186 | 0.20 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chrX_-_21776281 | 0.20 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chrX_-_14891150 | 0.20 |
ENST00000452869.1
ENST00000398334.1 ENST00000324138.3 |
FANCB
|
Fanconi anemia, complementation group B |
| chr12_+_49761147 | 0.20 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr15_-_78369994 | 0.19 |
ENST00000300584.3
ENST00000409931.3 |
TBC1D2B
|
TBC1 domain family, member 2B |
| chr18_+_72166564 | 0.19 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr11_-_62342375 | 0.19 |
ENST00000378019.3
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr4_+_76439649 | 0.18 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr6_+_36562132 | 0.18 |
ENST00000373715.6
ENST00000339436.7 |
SRSF3
|
serine/arginine-rich splicing factor 3 |
| chr17_+_38444115 | 0.18 |
ENST00000580824.1
ENST00000577249.1 |
CDC6
|
cell division cycle 6 |
| chr22_-_37584321 | 0.18 |
ENST00000397110.2
ENST00000337843.2 |
C1QTNF6
|
C1q and tumor necrosis factor related protein 6 |
| chr12_+_116985896 | 0.17 |
ENST00000547114.1
|
RP11-809C9.2
|
RP11-809C9.2 |
| chr5_+_138210919 | 0.17 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr11_-_46638720 | 0.17 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr1_-_26633067 | 0.17 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr22_+_45714672 | 0.16 |
ENST00000424557.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr7_+_56032652 | 0.16 |
ENST00000437587.1
|
GBAS
|
glioblastoma amplified sequence |
| chr10_-_70092671 | 0.16 |
ENST00000358769.2
ENST00000432941.1 ENST00000495025.2 |
PBLD
|
phenazine biosynthesis-like protein domain containing |
| chr1_-_165668100 | 0.16 |
ENST00000354775.4
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr3_+_124223586 | 0.16 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr1_+_174670143 | 0.16 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr19_+_15751689 | 0.16 |
ENST00000586182.2
ENST00000591058.1 ENST00000221307.8 |
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr9_+_116263778 | 0.15 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr12_+_48178706 | 0.15 |
ENST00000599515.1
|
AC004466.1
|
Uncharacterized protein |
| chr2_+_64069240 | 0.15 |
ENST00000497883.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr22_+_45714361 | 0.15 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr3_-_123512688 | 0.15 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr12_+_41136144 | 0.15 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr2_+_90458201 | 0.15 |
ENST00000603238.1
|
CH17-132F21.1
|
Uncharacterized protein |
| chr11_-_62607036 | 0.15 |
ENST00000311713.7
ENST00000278856.4 |
WDR74
|
WD repeat domain 74 |
| chr12_+_111051902 | 0.15 |
ENST00000397655.3
ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1
|
tectonic family member 1 |
| chr3_-_131753830 | 0.15 |
ENST00000429747.1
|
CPNE4
|
copine IV |
| chr1_+_65613340 | 0.15 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr2_+_204103733 | 0.15 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr5_+_162930114 | 0.14 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr19_+_14492247 | 0.14 |
ENST00000357355.3
ENST00000592261.2 ENST00000242786.5 |
CD97
|
CD97 molecule |
| chr2_+_132044330 | 0.14 |
ENST00000416266.1
|
CYP4F31P
|
cytochrome P450, family 4, subfamily F, polypeptide 31, pseudogene |
| chr6_-_111927062 | 0.14 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr2_-_172291273 | 0.14 |
ENST00000442778.1
ENST00000453846.1 |
METTL8
|
methyltransferase like 8 |
| chr5_-_159846066 | 0.14 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr14_+_36295638 | 0.14 |
ENST00000543183.1
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like |
| chr10_+_13203543 | 0.14 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chr1_-_68962805 | 0.14 |
ENST00000370966.5
|
DEPDC1
|
DEP domain containing 1 |
| chr1_+_174669653 | 0.14 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_49121165 | 0.14 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr1_+_158901329 | 0.13 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr7_-_67162002 | 0.13 |
ENST00000420758.1
|
RP4-736H5.3
|
RP4-736H5.3 |
| chr2_+_64073187 | 0.13 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr2_+_64069459 | 0.13 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr4_-_100212132 | 0.13 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr10_+_135207598 | 0.13 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr9_+_135906076 | 0.13 |
ENST00000372097.5
ENST00000440319.1 |
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa |
| chr2_+_161993465 | 0.13 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr4_-_104021009 | 0.13 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr14_-_90798273 | 0.13 |
ENST00000357904.3
|
NRDE2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr7_+_6071007 | 0.13 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr5_-_180688105 | 0.12 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr5_-_38557561 | 0.12 |
ENST00000511561.1
|
LIFR
|
leukemia inhibitory factor receptor alpha |
| chr11_+_35211429 | 0.12 |
ENST00000525688.1
ENST00000278385.6 ENST00000533222.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_120315074 | 0.12 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_+_168195229 | 0.12 |
ENST00000271375.4
ENST00000367825.3 |
SFT2D2
|
SFT2 domain containing 2 |
| chr1_+_109632425 | 0.12 |
ENST00000338272.8
|
TMEM167B
|
transmembrane protein 167B |
| chr4_-_46911223 | 0.12 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr15_+_43477580 | 0.12 |
ENST00000356633.5
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr2_-_38303218 | 0.12 |
ENST00000407341.1
ENST00000260630.3 |
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr22_+_25202232 | 0.12 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr15_+_67841330 | 0.12 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr17_-_41132410 | 0.11 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr11_+_120195992 | 0.11 |
ENST00000314475.2
ENST00000529187.1 |
TMEM136
|
transmembrane protein 136 |
| chr6_-_144329531 | 0.11 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr11_-_75017734 | 0.11 |
ENST00000532525.1
|
ARRB1
|
arrestin, beta 1 |
| chr3_+_113775576 | 0.11 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr12_+_79371565 | 0.11 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chr2_+_234590556 | 0.11 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr17_+_53345080 | 0.11 |
ENST00000572002.1
|
HLF
|
hepatic leukemia factor |
| chr12_+_111051832 | 0.11 |
ENST00000550703.2
ENST00000551590.1 |
TCTN1
|
tectonic family member 1 |
| chr10_+_99332529 | 0.11 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr7_-_86849883 | 0.11 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr2_-_54197915 | 0.11 |
ENST00000404125.1
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4 |
| chr10_-_13523073 | 0.11 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr5_+_156693091 | 0.10 |
ENST00000318218.6
ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_-_8059638 | 0.10 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr9_-_39239171 | 0.10 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr9_+_131447342 | 0.10 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr16_-_79804394 | 0.10 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr15_+_58702742 | 0.10 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr5_-_114631958 | 0.10 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr2_+_71357744 | 0.10 |
ENST00000498451.2
|
MPHOSPH10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr20_+_56964169 | 0.10 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr11_-_71753188 | 0.10 |
ENST00000543009.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_-_131478468 | 0.10 |
ENST00000536673.1
ENST00000542980.1 |
RP11-76C10.5
|
RP11-76C10.5 |
| chr17_+_75401152 | 0.10 |
ENST00000585930.1
|
SEPT9
|
septin 9 |
| chr17_+_53344945 | 0.10 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr12_+_56546363 | 0.10 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr2_+_64068844 | 0.10 |
ENST00000337130.5
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr1_-_240775447 | 0.10 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr14_-_69262947 | 0.10 |
ENST00000557086.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr18_-_33702078 | 0.10 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr12_-_122296755 | 0.10 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr3_-_47950745 | 0.10 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr13_-_45048386 | 0.10 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chrX_-_106362013 | 0.10 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr4_-_156297919 | 0.10 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr6_+_8652370 | 0.10 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr18_-_64271363 | 0.09 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr10_+_116697946 | 0.09 |
ENST00000298746.3
|
TRUB1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr10_+_27793197 | 0.09 |
ENST00000356940.6
ENST00000535776.1 |
RAB18
|
RAB18, member RAS oncogene family |
| chr11_+_28131821 | 0.09 |
ENST00000379199.2
ENST00000303459.6 |
METTL15
|
methyltransferase like 15 |
| chr4_-_84035905 | 0.09 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr7_-_38948774 | 0.09 |
ENST00000395969.2
ENST00000414632.1 ENST00000310301.4 |
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr7_+_1609765 | 0.09 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr1_+_202317855 | 0.09 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_-_104532062 | 0.09 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr21_-_46131470 | 0.09 |
ENST00000323084.4
|
TSPEAR
|
thrombospondin-type laminin G domain and EAR repeats |
| chr14_-_21516590 | 0.09 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr10_-_76868931 | 0.09 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr18_+_43684298 | 0.09 |
ENST00000282058.6
|
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr5_-_132200477 | 0.09 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr10_-_73976884 | 0.09 |
ENST00000317126.4
ENST00000545550.1 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr19_+_33865218 | 0.09 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr1_-_35325318 | 0.08 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
| chr14_+_55494323 | 0.08 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr4_-_84035868 | 0.08 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr14_-_45252031 | 0.08 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr3_-_122134882 | 0.08 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr5_-_54988448 | 0.08 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr19_+_14491948 | 0.08 |
ENST00000358600.3
|
CD97
|
CD97 molecule |
| chr12_-_122711968 | 0.08 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr4_-_141677267 | 0.08 |
ENST00000442267.2
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr1_+_230193521 | 0.08 |
ENST00000543760.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr4_+_146403912 | 0.08 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr9_-_99382065 | 0.08 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr7_+_134430212 | 0.08 |
ENST00000436461.2
|
CALD1
|
caldesmon 1 |
| chr16_-_2031464 | 0.08 |
ENST00000356120.4
ENST00000354249.4 |
NOXO1
|
NADPH oxidase organizer 1 |
| chr1_+_110091189 | 0.08 |
ENST00000369851.4
|
GNAI3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr4_+_128802016 | 0.08 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr3_+_186743261 | 0.08 |
ENST00000423451.1
ENST00000446170.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_+_46806452 | 0.07 |
ENST00000536062.1
|
NSUN4
|
NOP2/Sun domain family, member 4 |
| chrX_-_134049233 | 0.07 |
ENST00000370779.4
|
MOSPD1
|
motile sperm domain containing 1 |
| chr3_-_42623805 | 0.07 |
ENST00000456515.1
|
SEC22C
|
SEC22 vesicle trafficking protein homolog C (S. cerevisiae) |
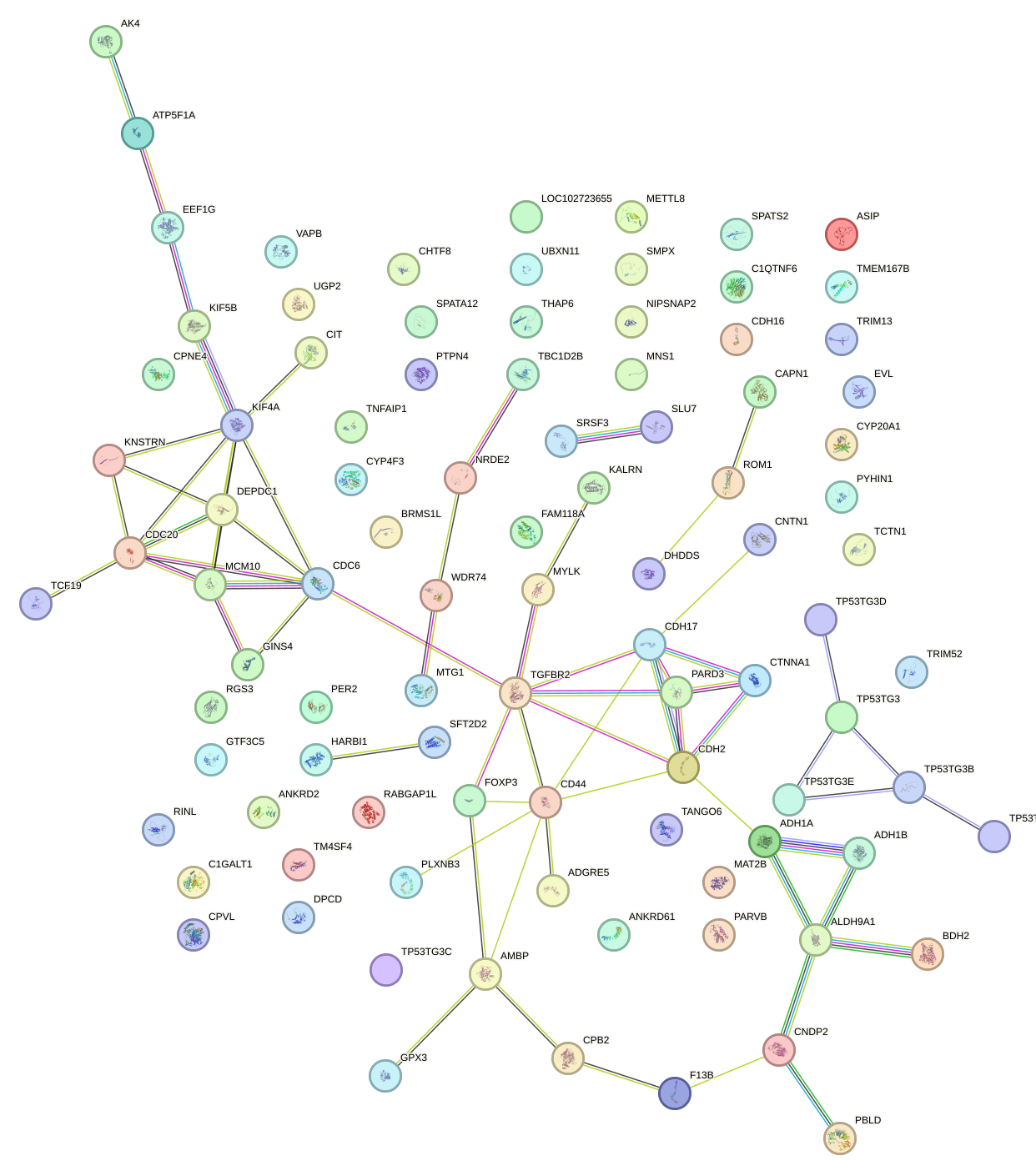
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.4 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.2 | GO:1902744 | negative regulation of lamellipodium assembly(GO:0010593) negative regulation of lamellipodium organization(GO:1902744) |
| 0.1 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:0002669 | tolerance induction dependent upon immune response(GO:0002461) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.2 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.3 | GO:2000809 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.4 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 1.4 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.5 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.5 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.1 | 0.4 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |