Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
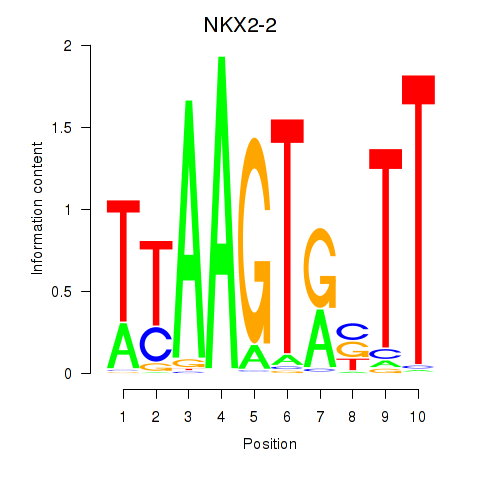
Results for NKX2-2
Z-value: 1.03
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NK2 homeobox 2 |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_30326445 | 0.93 |
ENST00000378963.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr12_-_102591604 | 0.75 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_+_241695424 | 0.73 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr11_-_8892900 | 0.71 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chrY_+_15418467 | 0.69 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr1_+_241695670 | 0.67 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr3_-_121379739 | 0.55 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr7_+_37723450 | 0.52 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr12_-_4488872 | 0.51 |
ENST00000237837.1
|
FGF23
|
fibroblast growth factor 23 |
| chr12_-_112614506 | 0.48 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr7_-_84121858 | 0.47 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr5_+_82767487 | 0.46 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr4_+_155484103 | 0.46 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr15_+_58702742 | 0.45 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr6_-_73935163 | 0.45 |
ENST00000370388.3
|
KHDC1L
|
KH homology domain containing 1-like |
| chr4_+_155484155 | 0.45 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr13_-_67802549 | 0.44 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr8_+_22414182 | 0.41 |
ENST00000524057.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr4_-_110723335 | 0.41 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr2_-_36779411 | 0.40 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr1_-_45253377 | 0.39 |
ENST00000372207.3
|
BEST4
|
bestrophin 4 |
| chr12_+_14572070 | 0.39 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr8_-_102181718 | 0.38 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr17_+_42015654 | 0.38 |
ENST00000565120.1
|
RP11-527L4.2
|
Uncharacterized protein |
| chr1_+_211433275 | 0.37 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr19_-_17366257 | 0.37 |
ENST00000594059.1
|
AC010646.3
|
Uncharacterized protein |
| chr21_+_17214724 | 0.37 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr15_-_38519066 | 0.37 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr12_-_120315074 | 0.36 |
ENST00000261833.7
ENST00000392521.2 |
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr12_-_42631529 | 0.35 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr9_-_97356075 | 0.35 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr17_+_15604513 | 0.35 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr18_+_3466248 | 0.34 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr5_+_82767583 | 0.34 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr1_+_104068312 | 0.33 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr8_+_71485681 | 0.33 |
ENST00000391684.1
|
AC120194.1
|
AC120194.1 |
| chr8_-_95220775 | 0.32 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr9_-_99382065 | 0.32 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr1_-_182573514 | 0.31 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr16_-_21289627 | 0.31 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr12_-_68696652 | 0.31 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr4_+_95679072 | 0.31 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr9_+_129097520 | 0.30 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr10_+_32735030 | 0.30 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr4_+_87928140 | 0.30 |
ENST00000307808.6
|
AFF1
|
AF4/FMR2 family, member 1 |
| chr7_-_120497178 | 0.30 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr1_-_207095324 | 0.29 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_+_115862858 | 0.29 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chrX_+_123095546 | 0.28 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr1_+_104068562 | 0.28 |
ENST00000423855.2
|
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr6_-_33285505 | 0.28 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr14_+_39734482 | 0.28 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr13_-_86373536 | 0.27 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr9_+_92219919 | 0.27 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr15_+_100348193 | 0.27 |
ENST00000558188.1
|
CTD-2054N24.2
|
Uncharacterized protein |
| chr13_-_107220455 | 0.27 |
ENST00000400198.3
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr18_-_68004529 | 0.27 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr17_+_75315654 | 0.26 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr3_+_189507432 | 0.26 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr4_+_80584903 | 0.26 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr12_-_371994 | 0.26 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr6_-_116833500 | 0.26 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr11_+_59705928 | 0.25 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr2_+_170655322 | 0.25 |
ENST00000260956.4
ENST00000417292.1 |
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr2_-_239198743 | 0.25 |
ENST00000440245.1
ENST00000431832.1 |
PER2
|
period circadian clock 2 |
| chr5_+_75904950 | 0.25 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr8_+_91233711 | 0.24 |
ENST00000523283.1
ENST00000517400.1 |
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr10_-_27529486 | 0.24 |
ENST00000375888.1
|
ACBD5
|
acyl-CoA binding domain containing 5 |
| chrX_+_123095860 | 0.23 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr22_-_43567750 | 0.23 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr1_+_53480598 | 0.23 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr1_+_171810606 | 0.23 |
ENST00000358155.4
ENST00000367733.2 ENST00000355305.5 ENST00000367731.1 |
DNM3
|
dynamin 3 |
| chr3_+_189507460 | 0.23 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr2_-_70409953 | 0.23 |
ENST00000419381.1
|
C2orf42
|
chromosome 2 open reading frame 42 |
| chrX_+_123095890 | 0.23 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr6_+_77484663 | 0.23 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr14_-_89878369 | 0.23 |
ENST00000553840.1
ENST00000556916.1 |
FOXN3
|
forkhead box N3 |
| chr2_+_189157536 | 0.22 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_-_111563076 | 0.22 |
ENST00000354925.2
ENST00000511990.1 |
PITX2
|
paired-like homeodomain 2 |
| chr21_+_17792672 | 0.22 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr9_-_39239171 | 0.22 |
ENST00000358144.2
|
CNTNAP3
|
contactin associated protein-like 3 |
| chr1_+_104159999 | 0.22 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr14_-_50154921 | 0.22 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr12_-_106697974 | 0.22 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr3_+_151591422 | 0.22 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr8_-_124749609 | 0.22 |
ENST00000262219.6
ENST00000419625.1 |
ANXA13
|
annexin A13 |
| chr4_+_128802016 | 0.22 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr10_-_112678692 | 0.21 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr7_+_23719749 | 0.21 |
ENST00000409192.3
ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A
|
family with sequence similarity 221, member A |
| chr2_-_75937994 | 0.21 |
ENST00000409857.3
ENST00000470503.1 ENST00000541687.1 ENST00000442309.1 |
GCFC2
|
GC-rich sequence DNA-binding factor 2 |
| chr4_+_74269956 | 0.21 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr2_-_75938115 | 0.21 |
ENST00000321027.3
|
GCFC2
|
GC-rich sequence DNA-binding factor 2 |
| chr3_+_98482175 | 0.21 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr1_-_75198940 | 0.21 |
ENST00000417775.1
|
CRYZ
|
crystallin, zeta (quinone reductase) |
| chr3_-_123339343 | 0.21 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr1_+_84630053 | 0.20 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_+_17135016 | 0.20 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr17_-_3496171 | 0.20 |
ENST00000399756.4
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr7_-_84569561 | 0.20 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr8_+_110552831 | 0.19 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr4_+_56719782 | 0.19 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr10_+_63808970 | 0.19 |
ENST00000309334.5
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr12_+_510795 | 0.19 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr9_-_85882145 | 0.19 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr1_-_26633480 | 0.19 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr5_+_35852797 | 0.19 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr9_+_91926103 | 0.19 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr21_+_30968360 | 0.18 |
ENST00000333765.4
|
GRIK1-AS2
|
GRIK1 antisense RNA 2 |
| chr3_-_122134882 | 0.18 |
ENST00000330689.4
|
WDR5B
|
WD repeat domain 5B |
| chr8_-_82644562 | 0.18 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr5_-_68339648 | 0.18 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr7_-_17598506 | 0.18 |
ENST00000451792.1
|
AC017060.1
|
AC017060.1 |
| chr1_-_115259337 | 0.18 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr15_+_36887069 | 0.17 |
ENST00000566807.1
ENST00000567389.1 ENST00000562877.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr4_-_178169927 | 0.17 |
ENST00000512516.1
|
RP11-487E13.1
|
Uncharacterized protein |
| chr12_+_510742 | 0.17 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr12_+_57998400 | 0.17 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chrX_+_108779004 | 0.17 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr2_+_39103103 | 0.17 |
ENST00000340556.6
ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2
|
MORN repeat containing 2 |
| chr18_+_43684298 | 0.17 |
ENST00000282058.6
|
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr13_+_25875662 | 0.17 |
ENST00000381736.3
ENST00000463407.1 ENST00000381718.3 |
NUPL1
|
nucleoporin like 1 |
| chr1_-_244006528 | 0.17 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr10_-_121296045 | 0.16 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr12_-_71551652 | 0.16 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr1_+_150898733 | 0.16 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chrX_-_70474499 | 0.16 |
ENST00000353904.2
|
ZMYM3
|
zinc finger, MYM-type 3 |
| chr5_+_159848854 | 0.16 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr2_+_113299990 | 0.16 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr5_+_159848807 | 0.16 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr1_-_160231451 | 0.16 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr14_-_82000140 | 0.15 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chrX_-_70474910 | 0.15 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr2_+_120436760 | 0.15 |
ENST00000445518.1
ENST00000409951.1 |
TMEM177
|
transmembrane protein 177 |
| chr18_+_19668021 | 0.15 |
ENST00000579830.1
|
RP11-595B24.2
|
Uncharacterized protein |
| chr17_-_29624343 | 0.15 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr20_-_13765526 | 0.15 |
ENST00000202816.1
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr3_+_189507523 | 0.15 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr2_+_183582774 | 0.15 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr4_+_41937131 | 0.14 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr12_+_52056548 | 0.14 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr13_+_50656307 | 0.14 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_+_67395922 | 0.14 |
ENST00000401042.3
ENST00000355356.3 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr12_-_111926342 | 0.14 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr15_+_56657613 | 0.14 |
ENST00000352903.2
ENST00000537232.1 ENST00000561221.2 ENST00000558083.2 |
TEX9
|
testis expressed 9 |
| chr2_+_162087577 | 0.14 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chrX_-_70473957 | 0.14 |
ENST00000373984.3
ENST00000314425.5 ENST00000373982.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr8_-_101719159 | 0.14 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_-_15510287 | 0.13 |
ENST00000397519.2
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr18_+_29671812 | 0.13 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr4_-_99578776 | 0.13 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr5_+_75904918 | 0.13 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr12_-_28124903 | 0.13 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chrX_-_70474377 | 0.13 |
ENST00000373978.1
ENST00000373981.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr14_-_100841930 | 0.13 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr1_+_61547894 | 0.13 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr4_+_174818390 | 0.13 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr1_-_246670614 | 0.13 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr2_-_55276320 | 0.13 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr8_-_133772870 | 0.13 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr5_+_148206156 | 0.12 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chrX_+_115567767 | 0.12 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr2_-_180871780 | 0.12 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr4_+_86699834 | 0.12 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_81601114 | 0.12 |
ENST00000602967.1
ENST00000429984.3 |
NUTM2E
|
NUT family member 2E |
| chr2_-_145275828 | 0.12 |
ENST00000392861.2
ENST00000409211.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_+_189157498 | 0.12 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_+_124320665 | 0.12 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr17_+_48823975 | 0.12 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr2_+_74229812 | 0.11 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr8_+_91233750 | 0.11 |
ENST00000523406.1
|
LINC00534
|
long intergenic non-protein coding RNA 534 |
| chr8_+_110552337 | 0.11 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr17_-_8059638 | 0.11 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr4_-_140477928 | 0.11 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_22222764 | 0.11 |
ENST00000439717.2
ENST00000412328.1 |
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr4_-_36246060 | 0.11 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr5_-_98262240 | 0.11 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr1_+_150898812 | 0.11 |
ENST00000271640.5
ENST00000448029.1 ENST00000368962.2 ENST00000534805.1 ENST00000368969.4 ENST00000368963.1 ENST00000498193.1 |
SETDB1
|
SET domain, bifurcated 1 |
| chr9_+_131447342 | 0.11 |
ENST00000409104.3
|
SET
|
SET nuclear oncogene |
| chr19_+_50148087 | 0.11 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr3_+_152017181 | 0.10 |
ENST00000498502.1
ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr3_-_57326704 | 0.10 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr17_+_48823896 | 0.10 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr11_-_26743546 | 0.10 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr8_+_76452097 | 0.10 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr12_-_8803128 | 0.10 |
ENST00000543467.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr10_+_81462983 | 0.10 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr6_-_27440460 | 0.10 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr12_-_10978957 | 0.10 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr3_+_75713481 | 0.10 |
ENST00000308062.3
ENST00000464571.1 |
FRG2C
|
FSHD region gene 2 family, member C |
| chr9_+_129097479 | 0.10 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr12_+_69202795 | 0.10 |
ENST00000539479.1
ENST00000393415.3 ENST00000523991.1 ENST00000543323.1 ENST00000393416.2 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr10_-_14504093 | 0.09 |
ENST00000493380.1
ENST00000475141.2 |
FRMD4A
|
FERM domain containing 4A |
| chr5_-_43557791 | 0.09 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr18_-_67615088 | 0.09 |
ENST00000582621.1
|
CD226
|
CD226 molecule |
| chr1_-_31538517 | 0.09 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr21_-_36421401 | 0.09 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr1_+_180601139 | 0.09 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr14_+_56584414 | 0.09 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_+_52978107 | 0.09 |
ENST00000445275.2
|
TOM1L1
|
target of myb1 (chicken)-like 1 |
| chr7_-_944631 | 0.09 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 0.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 0.5 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.5 | GO:0010966 | regulation of phosphate transport(GO:0010966) regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0019075 | virus maturation(GO:0019075) |
| 0.1 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.9 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0044727 | chromatin reprogramming in the zygote(GO:0044725) DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.3 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:2000323 | circadian regulation of translation(GO:0097167) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.3 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.9 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004583 | alpha-1,3-mannosyltransferase activity(GO:0000033) dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0004936 | adrenergic receptor activity(GO:0004935) alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |