Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
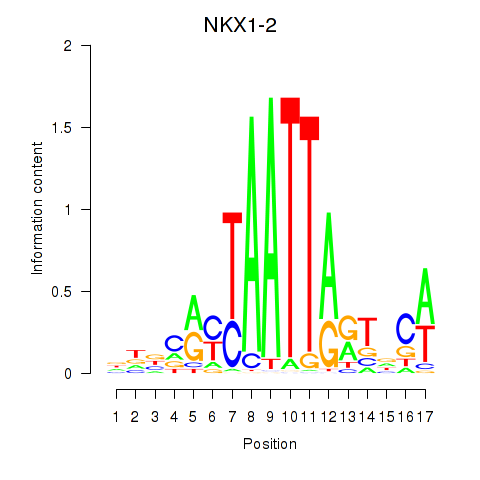
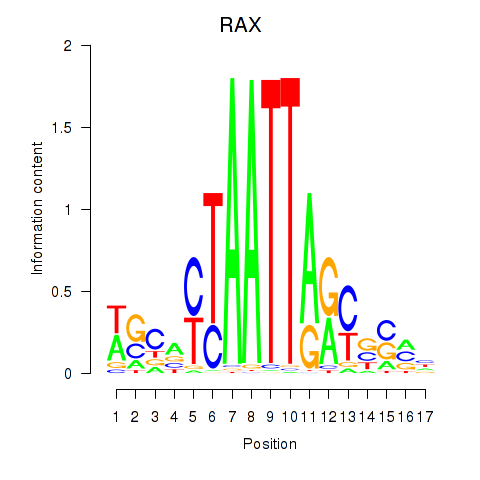
Results for NKX1-2_RAX
Z-value: 1.07
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-2
|
ENSG00000229544.6 | NK1 homeobox 2 |
|
RAX
|
ENSG00000134438.9 | retina and anterior neural fold homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAX | hg19_v2_chr18_-_56940611_56940660 | 0.96 | 4.3e-02 | Click! |
| NKX1-2 | hg19_v2_chr10_-_126138622_126138753 | 0.40 | 6.0e-01 | Click! |
Activity profile of NKX1-2_RAX motif
Sorted Z-values of NKX1-2_RAX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_57233087 | 2.13 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr17_-_57232525 | 1.55 |
ENST00000583380.1
ENST00000580541.1 ENST00000578105.1 ENST00000437036.2 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_89442940 | 1.53 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr17_-_57232596 | 1.34 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_+_57232690 | 1.32 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr8_-_101571964 | 0.91 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr10_+_99205959 | 0.88 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr3_+_158519654 | 0.85 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr10_+_99205894 | 0.78 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr4_+_66536248 | 0.78 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr2_+_13677795 | 0.75 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chrX_+_77166172 | 0.66 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr12_-_74686314 | 0.64 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr8_-_101571933 | 0.63 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr3_-_120400960 | 0.62 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr4_-_25865159 | 0.60 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr9_-_3469181 | 0.58 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr6_+_26440700 | 0.54 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chrX_+_43515467 | 0.53 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr14_-_53258314 | 0.53 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_+_138145076 | 0.53 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr17_-_45266542 | 0.52 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr1_+_28261492 | 0.50 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_201979645 | 0.49 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr15_+_96904487 | 0.49 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr2_+_27008865 | 0.46 |
ENST00000335756.4
ENST00000233505.8 |
CENPA
|
centromere protein A |
| chr8_-_102803163 | 0.44 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr17_-_4938712 | 0.43 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr17_-_27418537 | 0.41 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr16_+_53133070 | 0.37 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr9_-_5339873 | 0.35 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr2_-_239197238 | 0.35 |
ENST00000254657.3
|
PER2
|
period circadian clock 2 |
| chr8_+_107738240 | 0.33 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr2_-_239197201 | 0.33 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr9_+_135937365 | 0.31 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr13_+_78315295 | 0.31 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr12_-_112279694 | 0.31 |
ENST00000443596.1
ENST00000442119.1 |
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1 |
| chr1_+_162336686 | 0.30 |
ENST00000420220.1
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr6_+_130339710 | 0.29 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr19_+_8740061 | 0.28 |
ENST00000593792.1
|
CTD-2586B10.1
|
CTD-2586B10.1 |
| chr10_-_92681033 | 0.28 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr13_+_36050881 | 0.28 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr11_+_71900703 | 0.28 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr15_+_93443419 | 0.27 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr16_+_29832634 | 0.27 |
ENST00000565164.1
ENST00000570234.1 |
MVP
|
major vault protein |
| chr2_+_187371440 | 0.26 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr7_+_99717230 | 0.26 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr6_+_30687978 | 0.26 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr7_-_99716914 | 0.25 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_-_159832438 | 0.25 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr4_+_77356248 | 0.25 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr2_+_191221240 | 0.25 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr12_+_32832203 | 0.24 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr14_-_88200641 | 0.24 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr11_+_71900572 | 0.22 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_-_39341594 | 0.22 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr3_-_105587879 | 0.22 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_-_141348789 | 0.21 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr16_-_28608364 | 0.21 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr1_-_17766198 | 0.21 |
ENST00000375436.4
|
RCC2
|
regulator of chromosome condensation 2 |
| chr17_+_59489112 | 0.21 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr6_-_111927449 | 0.21 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr6_-_111927062 | 0.20 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr14_+_20187174 | 0.20 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr5_-_159546396 | 0.20 |
ENST00000523662.1
ENST00000456329.3 ENST00000307063.7 |
PWWP2A
|
PWWP domain containing 2A |
| chr6_+_52442083 | 0.20 |
ENST00000606714.1
|
TRAM2-AS1
|
TRAM2 antisense RNA 1 (head to head) |
| chr11_-_13011081 | 0.20 |
ENST00000532541.1
ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958
|
long intergenic non-protein coding RNA 958 |
| chr6_-_82957433 | 0.20 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_+_51851612 | 0.19 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr14_+_55494323 | 0.19 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr6_+_26365443 | 0.19 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr9_+_125133315 | 0.18 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_124329336 | 0.17 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr7_-_86849883 | 0.17 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr20_+_43990576 | 0.17 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr17_-_38821373 | 0.15 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr3_+_186353756 | 0.15 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr10_+_35894338 | 0.14 |
ENST00000321660.1
|
GJD4
|
gap junction protein, delta 4, 40.1kDa |
| chr4_-_103749105 | 0.14 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_28390637 | 0.14 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr7_+_21582638 | 0.14 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr16_-_28608424 | 0.14 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr14_+_55493920 | 0.13 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr12_+_16500599 | 0.13 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr20_-_50722183 | 0.13 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr8_+_107738343 | 0.13 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr17_-_33390667 | 0.13 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr16_-_28937027 | 0.12 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr19_+_37742773 | 0.11 |
ENST00000438770.2
ENST00000591116.1 ENST00000592712.1 |
AC012309.5
|
AC012309.5 |
| chr4_-_103749313 | 0.11 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_94890648 | 0.11 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr19_-_36822551 | 0.10 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr5_-_102898465 | 0.10 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr3_+_195447738 | 0.10 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr17_-_40337470 | 0.09 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr4_+_80584903 | 0.09 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr2_-_74618907 | 0.09 |
ENST00000421392.1
ENST00000437375.1 |
DCTN1
|
dynactin 1 |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr4_-_74486109 | 0.08 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_37488547 | 0.08 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr14_+_32798547 | 0.07 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr16_+_3333443 | 0.07 |
ENST00000572748.1
ENST00000573578.1 ENST00000574253.1 |
ZNF263
|
zinc finger protein 263 |
| chr5_+_66300446 | 0.07 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_36239576 | 0.07 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr12_+_16500571 | 0.07 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr2_-_190446738 | 0.07 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr11_+_118398178 | 0.07 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr6_+_160542870 | 0.07 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chrX_+_114827818 | 0.07 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr7_-_86849836 | 0.06 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chrX_+_77154935 | 0.06 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr8_-_30670384 | 0.05 |
ENST00000221138.4
ENST00000518243.1 |
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_-_12837423 | 0.05 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr12_+_7013897 | 0.05 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr22_-_32651326 | 0.05 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr1_+_153746683 | 0.04 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr2_+_24714729 | 0.04 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr14_+_67831576 | 0.04 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr21_-_35796241 | 0.04 |
ENST00000450895.1
|
AP000322.53
|
AP000322.53 |
| chr10_+_124913930 | 0.04 |
ENST00000368858.5
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr17_-_19364269 | 0.04 |
ENST00000421796.2
ENST00000585389.1 ENST00000609249.1 |
AC004448.5
|
AC004448.5 |
| chr10_+_124913793 | 0.04 |
ENST00000368865.4
ENST00000538238.1 ENST00000368859.2 |
BUB3
|
BUB3 mitotic checkpoint protein |
| chr1_+_186265399 | 0.04 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr17_+_39346139 | 0.03 |
ENST00000398470.1
ENST00000318329.5 |
KRTAP9-1
|
keratin associated protein 9-1 |
| chr8_-_29605625 | 0.03 |
ENST00000506121.3
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr10_+_124914285 | 0.03 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr11_+_29181503 | 0.03 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr9_+_109685630 | 0.03 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr12_+_93130311 | 0.03 |
ENST00000344636.3
|
PLEKHG7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr12_+_90674665 | 0.02 |
ENST00000549313.1
|
RP11-753N8.1
|
RP11-753N8.1 |
| chr4_+_110736659 | 0.02 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr17_+_1666108 | 0.02 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr8_+_9413410 | 0.02 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr3_-_180397256 | 0.01 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr2_+_217524323 | 0.01 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr1_-_159915386 | 0.01 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr4_-_66536196 | 0.01 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr7_-_87342564 | 0.01 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr2_+_112939365 | 0.01 |
ENST00000272559.4
|
FBLN7
|
fibulin 7 |
| chr10_+_6779326 | 0.01 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr2_-_198540719 | 0.01 |
ENST00000295049.4
|
RFTN2
|
raftlin family member 2 |
| chr11_+_112047087 | 0.01 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr1_-_151804314 | 0.01 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr14_+_74034310 | 0.00 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr4_-_103749205 | 0.00 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_100818009 | 0.00 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-2_RAX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.2 | 0.7 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.1 | 0.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.5 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.5 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 2.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0070839 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) divalent metal ion export(GO:0070839) |
| 0.0 | 0.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.4 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 2.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.2 | 0.7 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 3.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |